This is a default text for your page Sandbox2QRU. Click above on edit this page to modify. Be careful with the < and > signs.
The work below details the computational characterization and functional analysis of a protein, 2QRU, with a known crystal structure. Using a variety of computational methods, a variety of key identifiers in the sequences and structure could be isolated to better understand the function of the protein. This is due to the ideology that sequence denotes structure which denotes function.
2QRU is known to be a hydrolase, most active on esterase. The structure is 276 amino acids long, which is approximately 30.14 kDa. Knowing 2QRU is an alpha/beta hydrolase, the literature helped give further information on possible experimental conditions and substrates that can be used to identify activity. However, experimental data has not been obtained yet.
Structure/Sequence Analysis
Sequence Analysis
SPRITE
Analysis with Sprite displayed the proteins closest in structure to 2QRU. To do this, Sprite analyzed 2QRU for an active site with a motif of known catalytic function and compares to proteins known in the database. Among the proteins with the highest alignment (lowest RMSD) with a RMSD of 1.17 is 2LPR, a hydrolase. The amino acids included in the active site was 102 Ser, 104 Gly, 219 Asp, 247 His.
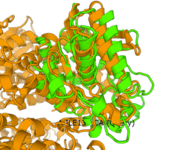
Chimera
Chimera allows for the visualization and structural comparison for 2QRU and similar proteins. Chimera uses the results gathered from the Sprite assay to display the proposed structure of 2QRU as well as that structure placed next to that of a similar protein. From the analysis, the structure of 2QRU most greatly resembles that of 1A7U with an RMSD value of 0.285 (compared to the next lowest value of 2.83).
BLAST
Blast is a sequence searching tool that allows for analysis of 2QRU’s protein sequence and therefore further predictions for the protein. The results display a list of proteins that are the closest in sequence to the one entered arranged by Query Coverage (higher value favored out of 100%), and E Value (lower value favored). Among the top Blast results for 2QRU many similarly structured proteins were listed under the family of alpha/beta hydrolases.
Structure Analysis
DALI
DALI analysis showed certain highly conserved sequences among certain related proteins.
Among all the highest related proteins, the 2YH2D showed the closest to the 2QRU active site. The 2QRU sequence has some similarity in short bursts, but nothing long-term. Two recurring sequences were HGGG at around 90 which was present a coil and SAGG around 175 which was present in a helical portion.
InterPRO
Investigation with InterPro showed the likely family of this enzyme was an alpha/beta hydrolase. Specifically, 2QRU was classified as a GDXG lipolytic enzyme. This family is generally associated with the hydrolysis of short fatty esters up to 8 carbons in length. Enzymes within this family are known to be involved in plant wall degradation and tryptophan synthesis.
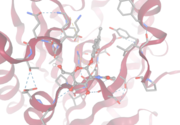
SwissDock
SwissDock was used to investigate the active site binding of different substrates.
The analysis of 2QRU on this site was performed with orientation parameters of 8x20x10 in a 25x25x25 Å box. The best overall interaction tested was with 4-Methylumbelliferyl 2-acetamido-2-deoxy-beta-D-glucopyranoside that showed an affinity -6.883 kcal/mol. Other significant binding affinities include 4-Nitrophenyl acetate at -5.093 kcal/mol and 4-Nitrophenyl acetate at -5.193 kcal/mol. The greatest binding affinities found were with carbohydrate-based substrates.
Proposed Functionality
Based on the sequence and structural analysis via computational modeling, it is known that the primary function of 2QRU is an alpha/beta hydrolase. Using the Swiss dock, we know the substrates with the highest bonding affinity were sugar-like molecules. This produces bond affinities of -6 or lower. Knowing this, it is hypothesized that 2QRU would be used in biological systems that need to break down sugars for energy, like fermentation or cellular respiration. Where exactly this would occur or on which sugar is unknown at this time, but experimental results may aid in finding what substrate works the best and in what environment.