This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
1fno
From Proteopedia
|
PEPTIDASE T (TRIPEPTIDASE)
Overview
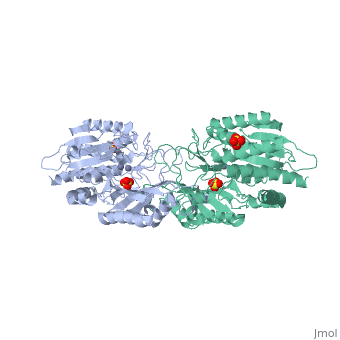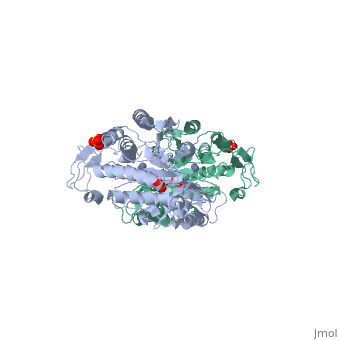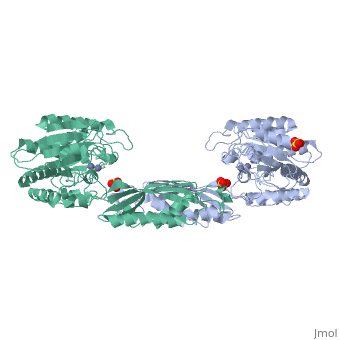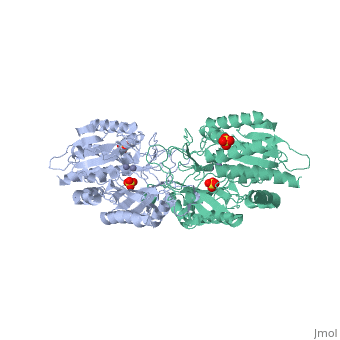
The structure of peptidase T, or tripeptidase, was determined by multiple, wavelength anomalous dispersion (MAD) methodology and refined to 2.4 A, resolution. Peptidase T comprises two domains; a catalytic domain with an, active site containing two metal ions, and a smaller domain formed through, a long insertion into the catalytic domain. The two metal ions, presumably, zinc, are separated by 3.3 A, and are coordinated by five carboxylate and, histidine ligands. The molecular surface of the active site is negatively, charged. Peptidase T has the same basic fold as carboxypeptidase G2. When, the structures of the two enzymes are superimposed, a number of homologous, residues, not evident from the sequence alone, could be identified., Comparison of the active sites of peptidase T, carboxypeptidase G2, Aeromonas proteolytica aminopeptidase, carboxypeptidase A and leucine, aminopeptidase reveals a common structural framework with interesting, similarities and differences in the active sites and in the zinc, coordination. A putative binding site for the C-terminal end of the, tripeptide substrate was found at a peptidase T specific fingerprint, sequence motif.
About this Structure
1FNO is a Single protein structure of sequence from Salmonella typhimurium with ZN and SO4 as ligands. Full crystallographic information is available from OCA.
Reference
Structure of peptidase T from Salmonella typhimurium., Hakansson K, Miller CG, Eur J Biochem. 2002 Jan;269(2):443-50. PMID:11856302
Page seeded by OCA on Tue Nov 20 15:04:13 2007