Antizyme Inhibitor
From Proteopedia
Crystal structure of the Antizyme inhibitor
| |||||||
| AzI, unpublished structure | |||||||
|---|---|---|---|---|---|---|---|
| Coordinates: | save as pdb, mmCIF, xml | ||||||
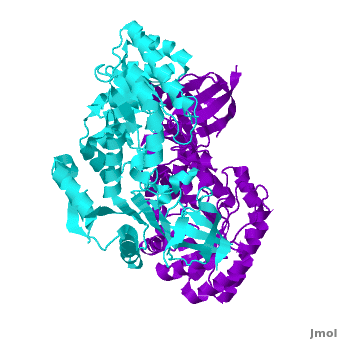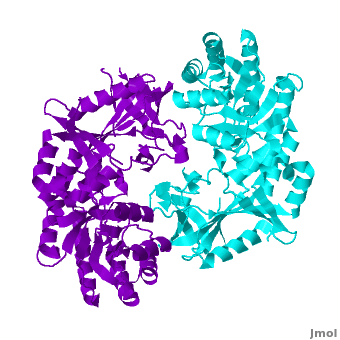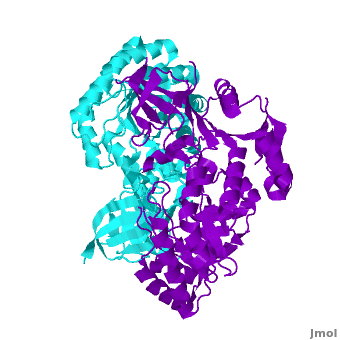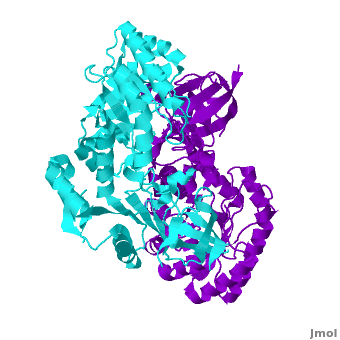
Antizyme inhibitor (AzI) regulates cellular polyamine homeostasis by binding to the polyamine-induced protein, Antizyme (Az), with greater affinity than ODC. AzI is highly homologous to ornithine decarboxylase (ODC), but is not enzymatically active. In order to understand these specific characteristics of AzI and its differences from ODC, we determined the 3D structure of mouse AzI to 2.05Å resolution. Both AzI and ODC crystallize as a (one monomer in cyan and the other in blue violet). However, fewer interactions at the dimer , a smaller buried surface area, and lack of symmetry of the interactions between residues from the two monomers in the AzI structure suggest that this dimeric structure is non-physiological. In addition, the absence of residues and interactions required for binding suggest that AzI does not bind PLP. A comparison to the revealed that AzI lacks the residues participating in PLP binding. Biochemical studies confirmed the lack of PLP binding and revealed that AzI exists as a monomer in solution while ODC is dimeric. Our findings that AzI exists as a monomer and its inability to bind PLP provide two independent explanations for its lack of enzymatic activity, and suggest the basis for its enhanced affinity towards Az.
|
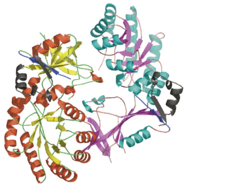
Each consists of two domains: a α/β-barrel [1] domain (residues 45–280) and a modified [2] β-sheet domain (residues 8–44 and 281–435). Helices are colored in red and β strands in yellow.
|
Comparison of the AzI structure to that of ODC. A primary sequence alignment and of mouse AzI crystallographic dimer (mAzI, cyan and blue violet) to mouse, human, and trypanosome ODC (mODC (PDB code 7odc, red and lime), hODC, and tODC, respectively) reveal high sequence identity (~50%) and structural similarity between AzI and ODC monomers (RMSD values of 1.85 Å, 1.6 Å, and 1.5 Å, respectively). Superposition of the of mAzI and mODC showing the variable loops between monomers A and B and between monomers A and B (AzI residues 355–362 and 387–401). AzI loops are in black, and ODC loops are in yellow. Structure of AzI suggests its existence as a monomer in solution. AzI crystallizes as a dimer such that the two monomers adopt a head-to-tail orientation as observed in the ODC dimer. The two monomers of AzI exhibit only (up 3.5 Å), while significantly more contacts are observed between the two monomers of hODC, mODC, and tODC (83, , and 69, respectively). Furthermore, the surface area buried by the two AzI monomers is smaller than that buried by the mODC monomers (1492 Å2 per monomer compared to 2284 Å2, which are 8% and 13% of the solvent accessible area, respectively). These properties support a very loose crystallographic dimer.
Primary Reference
Shira Albeck, Orly Dym, Tamar Unger, Zohar Snapir, Zippy Bercovich and Chaim Kahana. Crystallographic and biochemical studies revealing the structural basis for antizyme inhibitor function. Protein Sci. 2008 May; 17(5): 793-802. Epub 2008 Mar 27.
Proteopedia Page Contributors and Editors (what is this?)
Alexander Berchansky, Michal Harel, Joel L. Sussman, Dinesh Kulhary, David Canner, Jaime Prilusky, Orly Dym