1pgo
From Proteopedia
|
CRYSTALLOGRAPHIC STUDY OF COENZYME, COENZYME ANALOGUE AND SUBSTRATE BINDING IN 6-PHOSPHOGLUCONATE DEHYDROGENASE: IMPLICATIONS FOR NADP SPECIFICITY AND THE ENZYME MECHANISM
Overview
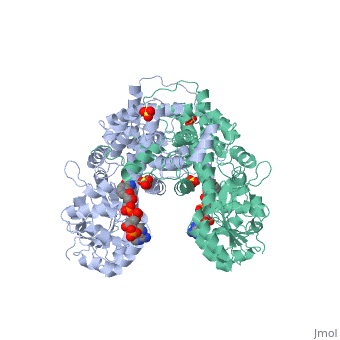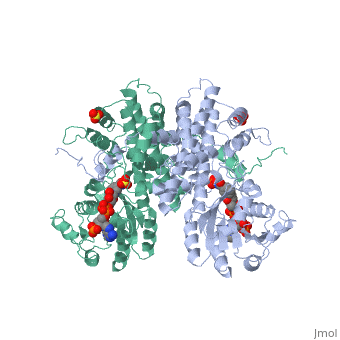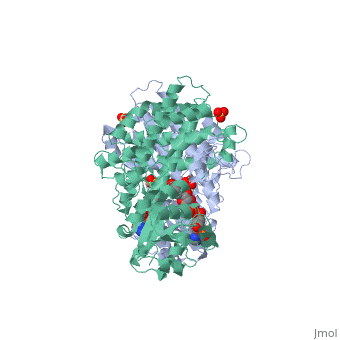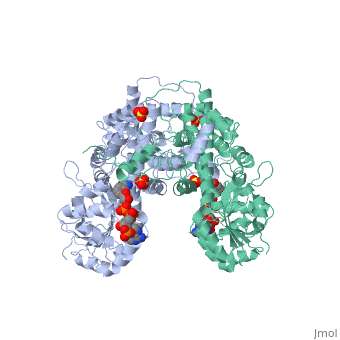
BACKGROUND: The nicotinamide adenine dinucleotide phosphate, (NADP)-dependent oxidative decarboxylase, 6-phosphogluconate, dehydrogenase, is a major source of reduced coenzyme for synthesis., Enzymes later in the pentose phosphate pathway convert the reaction, product, ribulose 5-phosphate, to ribose 5-phosphate. Crystallographic, study of complexes with coenzyme and substrate explain the NADP dependence, which determines the enzyme's metabolic role and support the proposed, general base-general acid mechanism. RESULTS: The refined structures of, binary coenzyme/analogue complexes show that Arg33 is ordered by binding, the 2'-phosphate, and provides one face of the adenine site. The, nicotinamide, while less tightly bound, is more extended when reduced than, when oxidized. All substrate binding residues are conserved; the, 3-hydroxyl of 6-phosphogluconate is hydrogen bonded to N zeta of Lys183, and the 3-hydrogen points towards the oxidized nicotinamide. The, 6-phosphate replaces a tightly bound sulphate in the apo-enzyme., CONCLUSIONS: NADP specificity is achieved primarily by Arg33 which binds, the 2'-phosphate but, in its absence, obscures the adenine pocket. The, bound oxidized nicotinamide is syn; hydride transfer from bound substrate, to the nicotinamide si- face is achieved with a small movement of the, nicotinamide nucleotide. Lys183 may act as general base. A water bound to, Gly130 in the coenzyme domain is the most likely acid required in, decarboxylation. The dihydronicotinamide ring of NADPH competes for, ligands with the 1-carboxyl of 6-phosphogluconate.
About this Structure
1PGO is a Single protein structure of sequence from Ovis aries with SO4 and NDP as ligands. Active as Phosphogluconate dehydrogenase (decarboxylating), with EC number 1.1.1.44 Full crystallographic information is available from OCA.
Reference
Crystallographic study of coenzyme, coenzyme analogue and substrate binding in 6-phosphogluconate dehydrogenase: implications for NADP specificity and the enzyme mechanism., Adams MJ, Ellis GH, Gover S, Naylor CE, Phillips C, Structure. 1994 Jul 15;2(7):651-68. PMID:7922042
Page seeded by OCA on Tue Nov 20 23:47:22 2007