We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Max Lein/Sandbox
From Proteopedia
| |||||||||
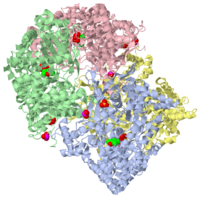
| 1gpa, resolution 2.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Non-Standard Residues: | |||||||||
| Activity: | Phosphorylase, with EC number 2.4.1.1 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
STRUCTURAL MECHANISM FOR GLYCOGEN PHOSPHORYLASE CONTROL BY PHOSPHORYLATION AND AMP
About this Structure
1GPA is a Single protein structure of sequence from Oryctolagus cuniculus. Additional information on 1GPA is available in a page on Glycogen Phosphorylase at the RCSB PDB Molecule of the Month. Full crystallographic information is available from OCA.
Reference
Structural mechanism for glycogen phosphorylase control by phosphorylation and AMP., Barford D, Hu SH, Johnson LN, J Mol Biol. 1991 Mar 5;218(1):233-60. PMID:1900534
Page seeded by OCA on Wed Jul 23 10:43:26 2008