Rebecca Martin/Sandbox1
From Proteopedia
Introduction to IgA
The most extensive surface in contact with the external environment is not our skin, but the epithelial lining of our gastrointestinal, respiratory, and urogenital tracts [1]. As a first line of defense in maintainance the integrity our mucosa, the immune system manufatures and secretes dimeric IgA to neutralize pathogenic organisms [2] and exclude the entry of commensals at the mucosal border [3]. In the serum, IgA functions as a second line of defense against pathogens that may breech the epithelial boundary [2]. The body produces more IgA than any other antibody isotype [3]. In fact, IgA is the most abundant antibody in the body, further illustrating IgA's critical role in immunity [4].
At least two isotypes exist, termed IgA1 and IgA2. IgA2 can further be categorized into 2 allotypes: IgA2 m(1) and IgA2 m(2). While IgA2 is found in most mammalian species, IgA1 is found only in higher apes. An approximately equal ratio of secretory IgA1 (sIgA1) to secretory IgA2 (sIgA2) reside at the mucosal surface, with the exception of the colon, where the majority is sIgA2 [5]. In the serum, about 90% of the IgA is monomeric IgA1 [4].
The receptors for IgA include the Fcα Receptor (FcαRI; CD89) and the polyimmunologlobulin receptor (pIgR). When binding to FcαRI results in the dimerization, the consequent signaling results in effector functions, including respiratory burst, mucosal surface, phaocytosis, and eosinophil degranulation. Binding to the pIgR results in transoocytosis and IgA secretion [2]. Unlike other antibody isotypes, IgA exists in mutiple oligomeric states [3]. The most common of which are the monomeric, dimeric, and secretory forms [4], adding to the complexity of structural functions for IgA. Exploring IgA's structure and protein interactions illuminates the unique and critical function IgA plays in humoral immunity.
Antibody Structure and the Immunoglobulin Domain
|
Overall Structure
- An antibody is a tetramer of and . In other words, the antibody is a of 2 heterodimers. Each is comprised on one light chain and one heavy chain. Heavy and light chains are held together with disulfide bonds and noncovalent interactions.
Fab and Fc fragments
- Another common way of describing antibody structure is in terms of its Fab and Fc fragments. Each light chains are composed of 2 immunoglobulin domains: one variable domain</scene> and one constant domain. Heavy chains composed of 4 Ig domains: one V-type and 3 C-type, named CH1 - CH3. A linking hinge region separates the CH2 and CH3 domains. Proteolytic cleavage at the hinge region by the protease papain, or a similar protease, yields 2 Fab fragments and 1 Fc fragment. Each contains 2 variable domains, one from the heavy chain and one from the light chain, and 2 constant domains one from the light chain and the Ch1 domain from the heavy chain. The Fc fragment contains 4 constant domains: the Ch2 and Ch3 domains from each of the heavy chains. Since the variable portions determine antigen specificity, the Fab fragments are generally thought of as the antigen-binding portion. The Fc fragment is important in binding various receptors, many of which are isotype specific and are named after the isotype of the ligand, i.e. FcαR binds the Fc portion of IgA.
Immunoglobulin domains
- The antibody is a member of the immunoglobulin superfamily of proteins (ref Att). Each chain can be further broken down into immunoglobulin domains: 2 in the light chain and 4 in the heavy chain, for a total of 12 in the entire antibody. Each immunoglobulin domain contains a primary amino acid sequence of approximately 70 – 100 residues long. Secondary structure is a characteristic beta sandwich with a variable number of beta strands, depending on the unit type. These strands display Greek key connectivity (web other) and form 2 beta sheets that fold over each other. An intra-domain disulfide bond stabilizes the tertiary structure.
-
- Nine antiparrallel beta strands comprise variable or V-regions. Loop sequences of varying length connect the strands. The 9 strands form 2 beta sheets, one with 4 (ABED-prosite) strands and the other with 3 (CFG prosite). The remaining 2 strands (C’ and C”) lie in between the 2 sheets. A disulfide bride stabilizes the 2 sandwich halves. Hydrophobic residues face the interior of the sheet, providing stability, while hydrophillic residues face outward and interact with the local environment. The extra loops in the V-region are critical for epitope specificity, and are consequently known as the compliment determining regions, here shown on the .
-
- C-type domains lack the C' and C beta strands. The sheets are ABED and CFG. Consequently, the sandich is more tightly packed. In the antibody, the constant domains determine the isotype: IgA, IgD, IgM, IgG, or IgE.
- Related structures
- Proteins containing the classic immunoglobulin-like domain are found predominantly in the immune system. In fact, the antibody's closest related structires are those that recognize antigen: MHC and TCRs.
- The V-type domain is found in a wider variety of proteins, including the Ig-binding molecules, such as the pIgR and the FcalphaR. Viral hemagluttinin is yet another example.
IgA1 and IgA2: a Structural COmparison
Hinge Region
- The hinge region differs significantly between the two IgA isoforms. The hinge region of IgA1 is comprised of 23 residues (PVPSTPPTPSPSTPPTPSPSCCH) and 5 O-glycosylation sites, while IgA2’s hinge region is comprised of 10 residues (PVPPPPPCCH) and no sites of glycosylation. Both hinge regions are located at Cys220 on the Ch1 chain and end at Ch2’s Pro244; however, the naming system is misleading, as it follows IgA1 and is therefore misleading. In fact, the distance from the the center of the 2 Fab fragments in IgA1 ia 16.9nm versus 8.2 nm in IgA2. So, while IgA1 remains extended, IgA2 is more compact. The greater number of residues in the IgA1 hinge region corresponds to a greater antigenic reach [2].
- These data must be taken into account with other hinge region characteristics. IgA1’s hinge region contains 5 sites of O-glycosylation, while IgA2’s hinge region contains none. In addition, IgA1’s hinge region contains 10 Pro residues, while IgA2’s region contains 6. In comparison, IgG’s hinge region contains No glycine residues reside in the hinge regions of either IgA1 or IgA2. The presence of prolines, the absence of glycine and the presence of glycosylated residues in IgA1 all amount to increased hinge rigidity in comparison to IgG1.
N-glycosylation
- In the harsh mucosal environment, glycosylated residues protect the protein from proteases. Both IgA1 and IgA2 display N-glycosylated residues. IgA1 has 3, at N263 on beta strand B on the Ch2 chain and on the J tail at N459. In IgA2, additional sites of N-glycosylation include Asn166 on the beta strand G of Ch1 and Asn337 of beta strand G on Ch2. Some alloforms of IgA2 are also N-glycosylated at Asn211 on Ch2. 15111057 An increased need for protection against proteolytic cleavage at the hinge region accounts for the presence of O-glycosylation in IgA1’s hinge region, particularly cleavage by bacterial metalloproteases. The glycosylation residues provide increased steric hindrance, and creating difficulty in fitting the peptide in the protease’s active site. In comparison to IgG, which is only 2.9% (w/w) glycosylated, IgA1 is 9.5% (w/w) and IgA2 is 11% (w/w) glycosylated. Overall, IgA1 is more susceptable to proteases than IgA2.
Disulfide Bonds
- The two structures also differ in the locations of their disulfide bonds 15111057 . In IgA1, a disulfide bond exists between the heavy chain Cys220 and light chain Cys196. This disulfide bond is absent in the main form of IgA2. Instead a disulfide bond links the 2 light chains at their C termini. The heavy and light chain associate through noncovalent interactions. So, while IgA1 may be more susceptable to proteases, IgA2 is more susceptable to denaturing conditions.
T-shape
- The unique characteristics of IgA1 and IgA2 explain the antibodys' overall T-shape. :IgA distinctly lacks the classic "Y-shape" antibody structure. IgA's increased hinge rigidity and a longer hinge region result in IgA1's predominately T-shape, in comparison to IgG's Y-shape. While the structure of IgA2 is more compact, the combination of an inter-light chain disulfide bond, a short hinge region, and proline residues with the hinge provide steric forces compatable with a T-shape. Of note, the T-shaped IgA2, with its interchain disulfide bond, resembles the structure of an IgG lacking the disulfide bonds between the heavy and light chains, which suggests the possibility of an evolutionary relationship between the two. The presence of IgA2 in lower mammals in contrast to IgA1 also supports this hypothesis.
Compare and Contrast
|
| ||||||
|---|---|---|---|---|---|---|---|
|
| ||||||
| IgM: Solution structure of human Immunoglobulin M 2rcj
IgG: Crystal structure of the intact human IgG B12 with broad and potent activity against primary HIV-1 isolates: a template for HIV vaccine design 1hzh IgG: Three=dimensional structure of a human immunoglobulin with a hinge deletion 1mco IgD: Semi-extended solution structure of human myeloma immunoglobulin D determined by constrained X-ray scattering 1zvo IgE: Structure of the human ige-fc bound to its high affinity receptor fc(epsilon)ri(alpha) 1f6a |
The J Chain allows IgA to form Dimers
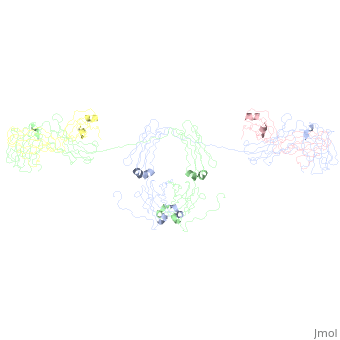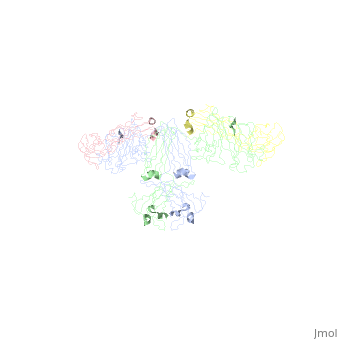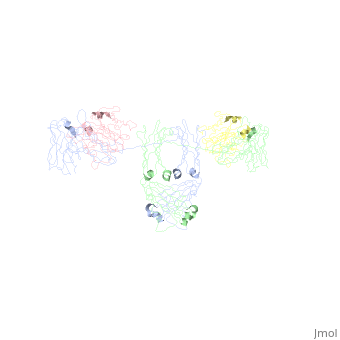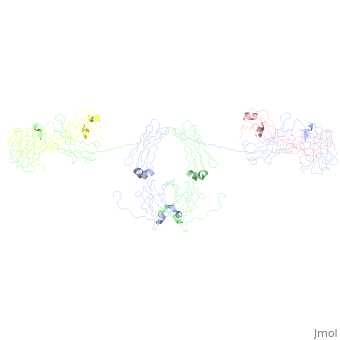
- The IgA structure has an addition 18 kDa, 137 residue polypeptide chain called the 10064707. This 18 kDa, 137-residue polypeptide chain is comprised of 2 immunoglobulin-like domains. The J chain is covalently attached to the C terminal Cys471 on IgA's Ch3 domain 18178841 via a disulfide bridge with either the J chain’s Cys 14 or the Cys 68. 10064707, 18178841 . The J chain has a single N-linked oligosaccharide 15111057, which increases rigidity and offers protection against proteases. The J chain allows IgA to form dimers, and less often trimer and tetramers. These polymers are rare because steric hindrance from the T-shaped Fab regions makes polymerization thermodynamically unfavorable 18178841.
|
- When IgA forms dimers, the Fc regions align end to end without overlap 18178841. The J chain lies within a fold in the bent Fc region. This conformation may allow the J chain access to the Secretory Component of the pIgR, which allows translocation across the mucosal epithelia to the luminal surface. Of note, in the image the J chains the J chains are extending from the dimer, which does not match with the described interaction of the J chain with the Fc portions of the anitbody (see Limitations of the Current Studies).
Secretory Component
- IgA is secreted as a dimer when it binds to the pIgR and is transported across the cell membrane 10064707 Upon IgA binding, the receptor-antibody complex is transocytosed to the lumenal side, where native proteases cleave the pIgR, releasing the secretory IgA (sIgA) into the lumen 10064707. The region of the pIgR that remains attached to the IgA upon pIgR cleavage is known as the .
The secretory compenent is the first 585 residues of the pIgR 17428798. The C terminal end of the secretory component is linked to the pIgR, but maintains no specific fold. The ability for the secretory to move freely facilitates its proteolytic cleavage and the secretion of sIgA. Structurally, the secretory component is comprised of 5 V-type immunoglobulin-like domains (D1-5) with 5-7 glycan chains, which increase the chains resistance to proteases. These glycosylation sites are located on one side of the protein and do not interfere with IgA binding. A long (10 amino acids) linker region exists between D3 and D4, so the D4 and D5 regions fold in on D2 and D3 in a compact J-shape. D1-3 are 12nm in length, while D4-5 are 10 nm long. Thus, D1 remains accessable. The one-sided glycans allow free access of D1's CDR regions and the Cys 502 at D5 to interact with IgA. It is thought that when D1 interacts with IgA's Fc region and the J chain, allowing the secretory component to unfold and disulfide formation between D5 C502 and IgA's Ch2 C311. While SC unfolds upon IgA binding, this binding imparts no change on the structure of IgA 19079336. Furthermore, the secretory component's binding site on IgA, interacting with the DE and FG loops affects the biding of IgA to its receptor FcalphaR, which interacts with the FG loops. 12768205 Fcalpha R, FG loops = major binding site for Fc 12768205 C311 only 10 angstroms away 12768205
Insights into Function
Glycosylation lends to protectiona against proteolytic attack in the harsh mucosal environment. Dimerization allows transcytosis and interferes with Fc receptor binding 4:1 --> 2:1 and then CS --> 1:1. COnsequently, it would be more difficult to elicit an immune response in the mucosa. Furthermore, unwanted inflammatory reactions to commensals would be more easilly avoided. Secretory Component results in unigue structure with IgA1 versus IgA2, acting in synergy and partaking in the antibody's antigen specificity. Since IgA1 is planar and more flexible, this might lend to antigen binding on proteins, which are larger and more variable. Flexibility allows IgA1 access to a more diverse array of orientations. Likewise the more compact, nonplanar IgA2 might. It is intersting to note that IgA2 tends to induce signaling at the Fca recetor. These synergistic relationships between structure and inter molecular interaciton suggest substantial coevolution between these molecules.
==synergy SC == mutual mucosal activity
changes in effector function
SIgA steric hindrance, no binding to mucosal surface 12768205 SIgA cannot activate Fcalpha R, FG loops = major binding site for Fc 12768205 C311 only 10 angstroms away 12768205 no binding to Fcalpha/ no activation w/o integrin 12768205 SC delays cleavage at Fc and hinge region, decreased access 2/2 fab and binding to cell surface R (bacterial proteases are alrge) 19079336 2 FcalphaRI binding sites, one per heavy chain, at each Ch2-Ch3 interface – both domains contribute 2:1 stoichiometry 15111057 Funct: helps prevent entry of pathogens/ gut flora into mucosa 10064707 SC steric hindrance pathogens cannot bind to mucosal surface 12768205 Function binding to IgA = protection against proteolytic degradation 17428798 Glycan residues do not impact binding aff to Iga, but instead assist with anchoring of the sIgA at the mucosal surface
Fc iga = more susc to intestinal proteases, precisely region pIg binds to and SC remains in assoc w Fc portion, w/o affecting function or motility of Fab or hinge region 17428798 Binding to Fc reduce flexibility @ hinge and btwn 2 Fc regions less lilkely to be in correct conformation for cleavage to occur 19079336
Implications in Science and Medicine
Proposed mechanism for IgA nephropathy: IgA nephropathy is the most prebvalent cause of chronic glomerulonephritis. This disease is caused by polymeric IgA1 deposited @ kidney glomeruli 18178841 Lack of the nepropathy in ppl w IgA myeloma w/o nephropathy suggest an abnormality in IgA structure. Notably, 90% of serum IgA is IgA1 and is monomeric. Propose disturbance in hinge region/ absence of fab (Steric hindrance of T-shaped fab regions polymers rare). Decreased O-glycosylation has been proposed as a mechanism- may destabilize hinge region, allow IgA to self associate or allow cleavage of hinge region by bacterial proteases. Conclusion: near-planar characteristic lends IgA1 to pathology 2/2 formation multimers following disruption of fab fragments from their natural rigid form Of note, studying mouse models of pathologies involving IgA1 may lack accuracy since IgA1 is found in higher apes only.
Limitations of the Current Studies
The techniques used in the majoroty of these studies were xray, neutron scattering analysis, analytical ultracentrifugation, and constrained modeling. Why didn't they just crystallize? Because IgA has a high amount of glycosylation and a relatiely large amount of flexibility, it has proven partivcularly difficult to crystalize in its intact form. 18178841, 10064707, 15111057 Glycosylation + linking regions btwn domains- longer = less likely to be crystallized 17428798 SC
Questions Unasnwered
Because of the limitating resolution of these models, many details concerning the binding residues and residue interactions are left unknown. SC aa interact w J chain? CDR-like motifs of SC's D1 bind where on @ IgA?; Locations of oligos on SC? Differences in binding IgA1 vs IgA2 17428798 Why does IgA2 lack as robust an effector function in binding to Fcalpha? Precise binding motifs SC and IgA1 18178841 Structure of IgA involved in IgA nephropathy 18178841 Crystallographic structure will yield further insights into the structure of IgA, the interactions between IgA and other molecules.
Links
IgA
- Fab and Fc Fragments
- Refined crystal structure of the galactan-binding immunoglobulin fab j539 at 1.95-angstroms resolution 2fbj
- Phosphocholine binding immunoglobulin fab mc/pc603. an x-ray diffraction study at 2.7 angstroms 1mcp
- Phosphocholine binding immunoglobulin fab mc/pc603. an x-ray diffraction study at 3.1 angstroms 2mcp
- Crystal structure of human FcaRI bound to IgA1-Fc 1ow0
- Refined crystal structure of a recombinant immunoglobulin domain and a complementarity-determining region 1-grafted mutant 2imm and2imn
- Crystal structure of a Staphylococcus aureus protein (SSL7) in complex with Fc of human IgA1 2qej
- Monomeric
- Dimeric and Secretory
Related Molecules
- non-IgA antibody isotypes
- IgM: Solution structure of human Immunoglobulin M 2rcj
- IgG: Crystal structure of the intact human IgG B12 with broad and potent activity against primary HIV-1 isolates: a template for HIV vaccine design 1hzh
- IgG: Three=dimensional structure of a human immunoglobulin with a hinge deletion 1mco
- IgD: Semi-extended solution structure of human myeloma immunoglobulin D determined by constrained X-ray scattering 1zvo
- IgE: Structure of the human ige-fc bound to its high affinity receptor fc(epsilon)ri(alpha) 1f6a
- Other C-type immunoglobulin examples
- V-type immunoglobulin examples
References
- ↑ Bonner A, Perrier C, Corthesy B, Perkins SJ. Solution structure of human secretory component and implications for biological function. J Biol Chem. 2007 Jun 8;282(23):16969-80. Epub 2007 Apr 11. PMID:17428798 doi:http://dx.doi.org/10.1074/jbc.M701281200
- ↑ 2.0 2.1 2.2 2.3 Furtado PB, Whitty PW, Robertson A, Eaton JT, Almogren A, Kerr MA, Woof JM, Perkins SJ. Solution structure determination of monomeric human IgA2 by X-ray and neutron scattering, analytical ultracentrifugation and constrained modelling: a comparison with monomeric human IgA1. J Mol Biol. 2004 May 14;338(5):921-41. PMID:15111057 doi:http://dx.doi.org/10.1016/j.jmb.2004.03.007
- ↑ 3.0 3.1 3.2 Bonner A, Almogren A, Furtado PB, Kerr MA, Perkins SJ. Location of secretory component on the Fc edge of dimeric IgA1 reveals insight into the role of secretory IgA1 in mucosal immunity. Mucosal Immunol. 2009 Jan;2(1):74-84. Epub 2008 Oct 8. PMID:19079336 doi:http://dx.doi.org/10.1038/mi.2008.68
- ↑ 4.0 4.1 4.2 Boehm MK, Woof JM, Kerr MA, Perkins SJ. The Fab and Fc fragments of IgA1 exhibit a different arrangement from that in IgG: a study by X-ray and neutron solution scattering and homology modelling. J Mol Biol. 1999 Mar 12;286(5):1421-47. PMID:10064707 doi:http://dx.doi.org/10.1006/jmbi.1998.2556
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namednineten