Off-Target Structural Insights: ArnA and AcrB in Bacterial Membrane Protein Cryo-EM Analysis
Mehmet Caliseki, Ufuk Borucu, Sathish K. N. Yadav, Christiane Schaffitzel
and Burak Veli Kabasakal [1]
Molecular Tour
Cells rely on finely tuned machinery to build and maintain their membranes, balancing the insertion, folding, and removal of proteins. In E. coli, this process is thought to involve the protease FtsH, the insertase YidC, and the regulatory HflKC complex. But their partnership had never been seen directly. To investigate, researchers turned to single-particle cryo-electron microscopy on detergent-solubilized samples enriched in these proteins.
What they found was surprising. Instead of the expected three-part assembly, the cryo-EM maps revealed exquisite structures of two other players: , an enzyme tied to polymyxin resistance, and , the major drug efflux transporter. of the final cryo-EM model (PDB ID 9v5r (blue)) with the reference crystal structure (PDB ID 7rr7, (yellow)) shows that they are remarkably similar, with an superposion of Cryo-EM vs crystal structure for AcrB with an RMSD between them, for CA atoms, of 2.0Å and
Both proteins are known to appear during affinity purification, but their repeated presence across different methods and even in membrane fractions suggests that their recovery was not purely accidental. ArnA, usually described as cytoplasmic, was consistently found in membrane-enriched samples, while AcrB is a well-established membrane protein. It was also observed that the class averages resembled GroEL and cytochrome bo3 oxidase.
These findings show that cryo-EM can capture not only the intended targets but also unexpected complexes that are well resolved and may have physiological importance. While only partial densities of the FtsH AAA+ domain were visible and no stable FtsH–YidC–HflKC complex could be reconstructed, the high-quality ArnA and AcrB structures provide fresh insights into bacterial survival strategies, from antibiotic resistance to drug efflux. More broadly, this study illustrates how structural biology can reveal unplanned discoveries that enrich our understanding of cell biology and the challenges of protein purification.
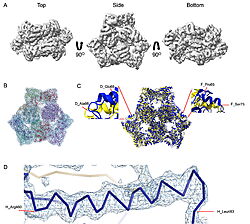 Cryo-EM density map and model fitting of the hexameric ArnA complex. (a) ArnA hexamer reconstructed at 4.0 Å resolution, shown from three orientations: top, side, and bottom views. The map reveals the characteristic two-layered architecture of ArnA and clearly resolved secondary-structure elements. (b) Final atomic model of ArnA fitted into the cryo-EM density map. Each of the 12 subunits is displayed in a different color to illustrate the hexameric arrangement. (c) Structural alignment of the final cryo-EM model (blue) (PDB ID 9y5h) with the reference crystal structure (yellow) (PDB ID 6pih) using ChimeraX. The alignment yielded an rmsd of 1.2 Å. Enlarged views show local conformational deviations in loop regions: Glu69–Ala98 in chain D (left) and Pro65–Ser75 in chain F (right). (d) Close-up view showing the model-to-map fit for a peptide segment (Leu483–Arg460). The density mesh is contoured at 1.7σ , showing clear peptide backbone density and supporting accurate model placement. 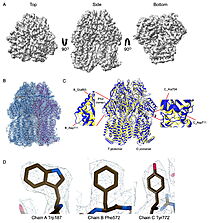 Cryo-EM density map and model fitting of the AcrB trimer. (a) AcrB reconstructed at 2.92 Å resolution, shown from top, side, and bottom. It reveals secondary-structure elements and transmembrane helices, consistent with the known trimeric architecture. (b) Final atomic model of AcrB fitted into the cryo-EM density map. Each protomer is shown in a different color to highlight the asymmetric trimer organization. (c) Alignment of the cryo-EM model (blue)(PDB-ID 9v5r) with the reference crystal structure, (yellow) (PDB ID 7rr7), performed using ChimeraX. The rmsd between the structures is 1.2 Å. Enlarged views highlight conformational deviations, including E693–D711 in protomer B (left) and A704–D711 in protomer C (right), located within the PN2 subdomain. (d) View of the density fit for (W187, chain A, F572, chain B, W772. chain C). The mesh is contoured at 1.6σ in Coot, showing well-defined density for aromatic residues. |
References
- ↑ doi: https://dx.doi.org/10.1107/S2059798325007089



