Journal:Acta Cryst F:S2053230X21013455
From Proteopedia

Crystal structure of betaine aldehyde dehydrogenase from Burkholderia pseudomalleiDylan K. Beard, Sandhya Subramanian, Jan Abendroth, Thomas E. Edwards, Peter J. Myler, and Oluwatoyin A. Asojo [1] Molecular Tour
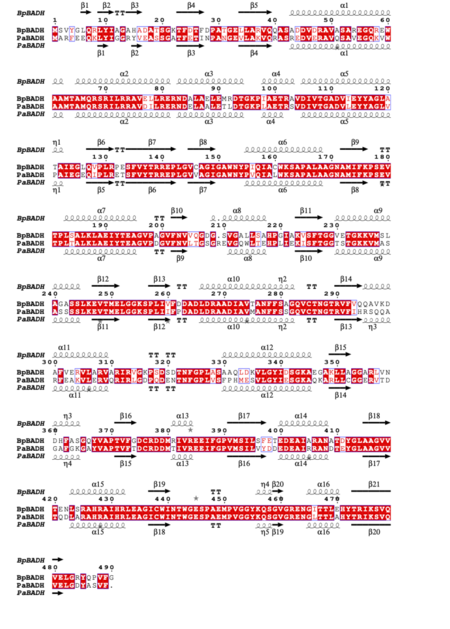
BpBADH has a prototypical BADH topology and shares considerable structure and sequence similarity with the ortholog from P. aeruginosa (PaBADH); see static image below: The structures are similar to those of BADH from Pseudomonas aeruginosa (PaBADH). The co-factor binding domains of BpBADH (6wsb) and PaBADH (4caz) are well conserved (identical residues of both structures are labeled in green, while non-identical in red): PaBADH is inhibited by the drug disulfiram which is an approved drug. Both structures show the irreversibly inhibited by disulfiram. Our preliminary analysis could facilitate drug repurposing studies for melioidosis. This project is an educational collaboration between the SSGCID and Hampton University. The SSGCID consortium is directed by Dr. Peter Myler (principal investigator) and comprises many different scientists working at multiple centers towards determining the three-dimensional structures of proteins from biodefense organisms and emerging infectious diseases. Dylan K. Beard was part of a pilot Hampton University Chemistry Education and Mentorship Course-based undergraduate research (HU-ChEM CURES) funded by the NIGMS. PDB references: betaine aldehyde dehydrogenase, 6wsa; bound to cofactor, 6wsb. References
| |||||||||||