ClpP Introduction
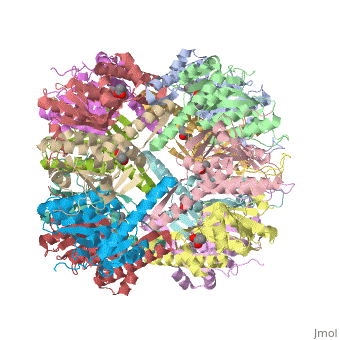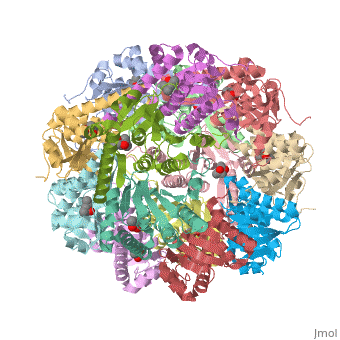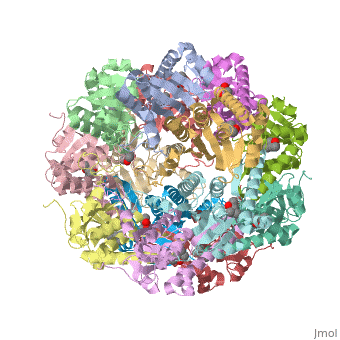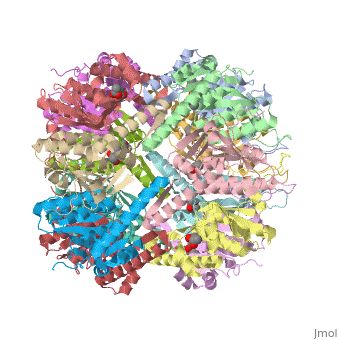
E. coli Casein lytic proteinase P (ClpP) is a double ring tetradecameric homo oligomer compartmentalized peptidase [1]. ClpP requires the use of ATP dependent regulatory elements that independently bind to ClpP in order for substrates to have access the active core [2][3][4]. Here, proteins that are translocated by regulatory elements into the peptidase core are cleaved into smaller amino acid chains approximately on average 6-8aa in length [5].
Tetradecameric Structure
ClpP is a serine protease which consists of fourteen monomers situated into two heptameric rings seated on top of each other. In the center of the barrel-shaped chamber lies a core of fourteen peptide-cleaving active sites, restricted by the narrow entrance called the axial pore. As peptides are processively threaded in the pore by the regulatory elements, the peptides are then degraded into smaller fragments as a result from ClpP cleavage. Smaller peptides are then released through small openings found around the equatorial interface of the two stacked rings.
Structural highlights
Featured within the core of ClpP are the catalytic sets of amino acids [X-Y-Z] that constitute the active mechanism featured by serene proteases [6][7]. Human ClpP can exist in two forms, a single heptameric ring or as a double stack set of hepatmeric rings where the double stacked form (tetradecamer) is the active form [8]. Tetradecamer ClpP is a stable but not a rigid structure that can undergo several conformational forms when interacting with its regulatory elements. By doing so both stabilizes processivity of translocation by the regulatory elements and is thought to increase likelihood of exposure to the active sites resulting in timely, small peptide formation.
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
Role of ClpP/Biological Relevance
E. coli ClpAP/ClpXP complexes play a critical role in maintaining protein homeostasis under several levels of quality control. Improperly folded or aggregated proteins are potential ClpP substrates based on properties of the associated regulatory element recognition. Targeted removal of aberrant proteins resulting from and rescue of stalled ribosomes by the SsrA tagging system are directly recognized and degraded by ClpAP/ClpXP complexes [9]. In E. coli ClpP and ClpP homologues found in other bacteria require regulatory elements to recognize and import proteins for destruction. To gain access to the active sites is tightly controlled and therefore a potential antimicrobial target where loss of regulation (for example, through use of acyldepsipeptides or ADEPs) literally digests the bacteria from the inside out [10]. E. coli ClpAP and ClpXP has been used as a structural model for the 26 proteasome to gain insight into its workings [11][12].