Sandbox 51
From Proteopedia
| Please do NOT make changes to this Sandbox. Sandboxes 30-60 are reserved for use by Biochemistry 410 & 412 at Messiah College taught by Dr. Hannah Tims during Fall 2012 and Spring 2013. |
|
Contents |
Introduction
Pancreatic lipase 1hpl (EC 3.1.1.3) is a an enzyme involved with the digestion and absorption of triacylglycerols (fats) in the intestine. It is secreted by the pancreas into the duodenum where it participates in the initial stages of breaking down fats into glyercol and three fatty acids. Lipase is a serine protease and a polar molecule. The mechanism of action requires Calcium ligands for stability and the coenzyme, colipase, for activation of the lipase mechanism. The hydrolysis products diffuse across enterocyte membrane and are absorbed into the blood [1]. Due to that role lipase has in digestion of fat, and the growing problem of obesity, there has been increased effort to develop lipase inhibitors for weight loss supplements.
Structure
1hpl is the horse pancreatic lipase enzyme that is thought to have a similar structure and function to the human pancreatic lipase. The of the human pancreatic lipase has not yet been published. Lipase is a dimer of two with 449 amino acid residues interacting with one each. The calcium ion shows that the molecule is located between acidic residues Arg, Asp and Glu. The enzyme has of various composition specific for certain interactions, an N-terminal (blue) and a C-terminal (red). In the , the N-terminal domain has the hydrolase alpha/beta folding structure, consisting of an 8 stranded alpha-beta sheet connected by helices. The C-terminal domain (where enzyme colipase binds) has a beta-sheet sandwich folding pattern [2]. The enzyme has 13 alpha helices (pink) and 22 beta sheets (yellow) per subunit, as displayed in the [3]. At the of the two monomers, interactions include 4 hydrogen bonds and 4 salt bridges that stabilize the dimer [4]. The molecule has a varying degree of interspersed within the protein(polar are shown in purple and hydrophobic in white) [5]. This is important because the enzyme actively digests at the lipid-water interface of the fatty micelles, requiring stability in both polar and non polar environments [6]. Within each monomer and dimer structure, the molecules are held together by disulfide bonds, hydrogen bonding, and electrostatic interactions (salt bridges). The enzyme has six covalent per monomer. Also, the stabilize the monomers and dimer of the enzyme at positively charged nitrogens (blue) in Arg and Lys, and negative oxygens (red) in Asp and Glu residues. are present within each monomer, as shown by the hydrogen bond forming residues (light gray), and the oxygen (red) and nitrogen (blue) atoms involved in the hydrogen bonding. Lipase is water soluble due to the polar residues on the surface, but has hydrophobic sequences on the interior. At enzyme activation and interaction with colipase, a confirmation change occurs to expose the more hydrophobic regions to the nonpolar lipid micelle.
Active Site
The of the lipase molecule is found in the N-terminal domain (residues 1-336) and contains a consisting of a Ser152-His263-Arg176 for the ester hydrolysis reaction (similar to that of a serine protease). The catalytic triad and regions around it are thought to be the best conserved aspects of lipase throughout the lipase family. The active site is covered by a 25-residue helical 'lid' blocking the binding site [7]. in complex with a triacylglyceride shows the unique lid (yellow) and beta-5 loop (pink) essential to catalysis. The 10 residue beta-5 loop changes confirmation when colipase binds, exposing lipase's oxyanion hole and hydrophobic surface. Before colipase binding however, lipase is in the structure where the beta-5 loop (residues 76-84, in pink) and lid (residues 237-261, yellow) protect the oxyanion hole from solvent interaction. The structure is accompanied by peptide shift which increase the hydrophobic surface area initiating the reaction with the lipid. one such important shift is at residues 240-253 (in yellow), in the lid structure, very close to the active site. Also, it is evident from the image that part of the beta-5 loop interacts with colipase in the open state. The lid opening is accompanied by a change in secondary structure from a mostly beta-extended confirmation to a structure where more than half the active site is formed from alpha helices [8].
 [9].
[9].
Function
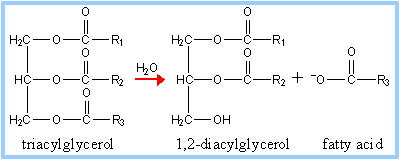
Pancreatic lipase initiates the breakdown of triacylglycerols into 1,2-diacylglycerols and fatty acids. Lipase is essential to hydrolyzing the very nonpolar triacylglycerols into more polar compounds capable of diffusing across enterocyte membranes, and eventually into circulation. The digestion of triacylglycerols is a major source of energy storage in metabolism. Consequently, lipase hydrolyzes the ester bonds of the triglycerides. Lipase, as a digestive enzyme, is soluble in water but the triacylglycerides it hydrolyzes are not. In the duodenum, bile salts initiate lipase's mechanism by emulsification of the fatty acids. Bile salts are usually sodium with the deprontated form of digestive acids (mostly derivatives of cholic acid). Bile salts are stored by the gallbladder and released into the duodenum where they facilitate the formation of micelles. Bile salts or acids are amphipathic. Therefore, the hydrophobic region surrounds the fatty acids and the hydrophilic remains on the outside in the process of emulsification, creating smaller micelle molecules. Micelle formation is essential to maximize the efficiency of lipase by increasing the hydrophilic surface area on which lipase will work [11]. For the hydrolysis reaction, lipase activity requires bile acids, its coenzyme (colipase), and calcium ions to achieve the correct orientation to hydrolyze the fats [12].
|
Mechanism
In addition to the effects of bile salts, Lipase is activated by the coenzyme colipase, which binds to the C-terminal non-catalytic domain of lipase. Upon binding, active lipase is stabilized for the hydrophobic interaction with the triacylglycerides [13]. Colipase must be present for activation of lipase and acts as a bridge between lipase and the lipid [14]. Colipase is also secreted in the pancreas, but in its inactive form, which must be activated by trypsin before interacting with lipase. Colipase is a small protein cofactor with 5 conserved disulfide bonds [15], and 2 surfaces- a hydrophilic surface (site of lipase C-terminal interaction) and a hydrophobic surface (contains multiple hydrophobic loops to bridge the lipid)[16]. Colipase and lipase are opposite of the active site on the C-terminal (contacts are regions of pink and yellow, with water molecules shown in darker blue). The enzymes are bound by polar interactions such as and van der waals forces [17]. Specifically, interactions at the include bonding between residues Glu15 and Arg38 with the lipase polar lid; and Arg44, Glu45, Glu64, Arg65, and Asn89 residues with main chain, C-terminal lipase residues [18].
Lipase activation at the lipid-water interface of triacylglycerides, in the presence of colipase and bile salts, is known as interfacial activation. For the hydroloysis reaction to take place, colipase anchors lipase to the lipid-water membrane of the micelle and a surface change occurs on lipase. Colipase's 4 hydrophobic loops interact with the hydrophobic atmosphere of the triacylglyceride initiating the lipase active site binding to the lipid, and lid opening to reveal a more hydrophobic environment for the triacylglycerol. Once colipase is bound, lipase initiates a serine-like hydrolysis involving the His-Asp-Ser active site residues in the catalytic triad [19]. In the reaction, serine attacks the ester, forming an acyl-enzyme intermediate. The His and Asp residues help to stabilize the oxyanion intermediate through hydrogen bonding. Water enters the active site and reacts to release lipase and free the fatty acid. This acylation and deacylation reaction is usually completely reversible.
Lipase Triacylglyceride Hydrolysis Mechanism
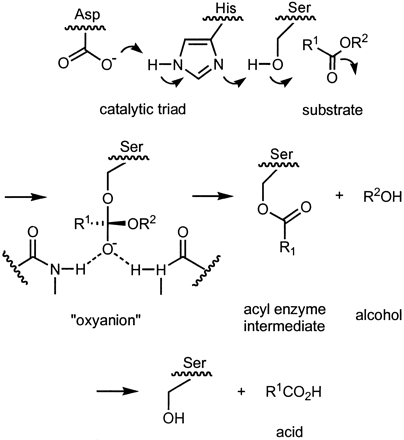 [20].
[20].
|
Inhibition
The inhibition of pancreatic lipase has serious effects on storage and absorption of fats taken in by the body, and is therefore a potentially strong basis for pharmaceuticals to combat obesity. Because lipase is a part of the serine esterase family, it is inhibited in a similar manner. One such compound is a inhibitor (shown interacting with the lipase-coplipase structure). The inhibitor acts by binding in the active site inducing conformational changes in the beta-5 loop structures. The alkyl chain fits into the hydrophobic portion of the active lipase-colipase complex mimicking the fatty acid produced through hydrolysis in the normal enzymatic reaction [21]. Van der waals forces between hydrophobic residues (blue) in the groove stabilize C11P binding, in addition to salt bridging and Hydrogen bonding forces with a cluster of hydrophilic residues (pick) around the Ser152 residue (purple) in the . [22]. Due to lipase's activity in the digestion and absorption of fat, there has been a growing market for lipase inhibitors for weight loss pharmaceuticals. The most popular is Orlistat (or Xenical®) which is a natural product from Streptomyces toxytricini and is the hydrogenation product of lipostation- an irreversible lipase inhibitor. This inhibitor also acts by binding Ser152, producing an ester which hydrolyzes so slow that it is practically irreversible [23].
References
- ↑ Voet, D.,etc. "Fundamentals of Biochemistry: Life at the Molecular Level" John Wiley and Sons, Inc: New Jersey, 2008.
- ↑ Horse pancreatic lipase...
- ↑ Egloff, M.P., etc. "The 2.46 angstroms resolution structure of the pancreatic lipase colipase complex inhibited by a C11 alkyl phosphonate."(1995) J. Biochemistry 34: 2751-2762 [1]
- ↑ "HORSE PANCREATIC LIPASE. THE CRYSTAL STRUCTURE AT 2.3 ANGSTROMS RESOLUTION." http://bisc.cse.ucsc.edu/pages/BiscHom/1hpl_AB.html
- ↑ Bourne, Y., etc. "Horse pancreatic lipase..."(1994) J.Mol.Biol. 238: 709-732 [2]
- ↑ Fundamentals of Biochemistry...
- ↑ Fundamentals of Biochemistry...
- ↑ Thomas, A. etc. "Role of the Lid Hydrophobicity Pattern in Pancreatic Lipase Activity", The Journal of Biological Chemistry, 2005 September 22; 270 (48): 40074-40083.
- ↑ "Role of the Lid Hydrophobicity Pattern in Pancreatic Lipase Activity"...
- ↑ http://www.rpi.edu/dept/bcbp/molbiochem/MBWeb/mb2/part1/lipoprot.htm
- ↑ Bowen, R. Absorption of Lipids. 8 August 2007. 3 March 2012. <http://www.vivo.colostate.edu/hbooks/pathphys/digestion/smallgut/absorb_lipids.html>.
- ↑ Fundamentals of Biochemistry...
- ↑ Fundamentals of Biochemistry...
- ↑ Crandall,W., Lowe, M. "Colipase Residues Glu64 and Arg65 Are Essential for Normal Lipase-mediated Fat Digestion in the Presence of Bile Salt Micelles" Journal of Biological Chemistry, 2001, (276) 12505-12512
- ↑ "Colipase". Wikipedia: The Free Encyclopedia. 5 July 2011 [3]
- ↑ "Colipase Residues..."
- ↑ van Tilbeurgh H, etc."Structure of the pancreatic lipase-procolipase complex", 1992 Sep 10;359(6391):159-62. PMID:1522902.[4]
- ↑ "Colipase Residues..."
- ↑ Fundamentals of Biochemistry...
- ↑ Reetz, Manfield F. Controlling the enantioselectivity of enzymes by directed evolution: Practical and theoretical ramifications. PNAS: 12 April 2004 [5]
- ↑ Egloff, M.P., etc. "The 2.46 angstroms resolution structure of the pancreatic lipase colipase complex inhibited by a C11 alkyl phosphonate."(1995) J. Biochemistry 34: 2751-2762 [6]
- ↑ Egloff, M., Marguet, F., Buono, G.,Verger,R.,Cambillau,C., Tilbeurgho,H. The 2.46 A Resolution Structure of the Pancreatic Lipase-Colipase Complex Inhibited by a C11 Alkyl Phosphonate? Biochemistry, 1995, 34, 275 1-2762. http://pubs.acs.org.ezproxy.messiah.edu/doi/pdf/10.1021/bi00009a003
- ↑ Kordik, C., Reitz, A. "Pharmacological Treatment of Obesity: Therapeutic Strategies" Journal of Medicinal Chemistry, 1999 (42).