User:Eileen Molzberger/Sandbox 1
From Proteopedia
Contents |
RecA
Introduction
RecA is a protein involved in several processes in a cell. These processes include DNA repair, and signalling the cell that DNA damage has occurred (formally referred to as SOS response induction). A ubiquitous protein, RecA or a RecA homolog is present in almost all cells.RecA Function in DNA Repair
DNA damage is a frequent occurrence. It can arise spontaneously, or can be caused other sources; these sources include reactive oxygen species formed during normal metabolic processes, Ultraviolet light and Ionizing Radiation. DNA damage that is not repaired can impair cell function or trigger programmed cell death (formally called "apoptosis"). DNA is composed to two complementary strands. When one of these strands is damaged, the cell can use the other undamaged strand to fill in the missing information on the damaged strand. Several different pathways exist to repair DNA, and RecA is involved in a pathway called recombinational repair. This pathway is one that uses the complementary strand to repair a damaged strand in a complex reaction called "strand exchange."
This process is especially important during DNA replication. During DNA replication, the two original DNA strands are separated and new, complementary strands are added to each of the original strands. The result is two new sets of double-stranded DNA (sometimes referred to as “duplexes”) exactly identical to the original double-stranded DNA set. So no unpaired DNA is present in either of the two new sets of DNA. A problem arises if one of the original strands is damaged; DNA damage is like a road block for the DNA replication machinery. If the original strand is damaged, a portion of that DNA strand won't be replicated. This results in a gap on the newly replicated strand. This gap is an unpaired segment of single-stranded DNA. Unpaired, single-stranded DNA is a red flag for the cell that something has gone wrong!
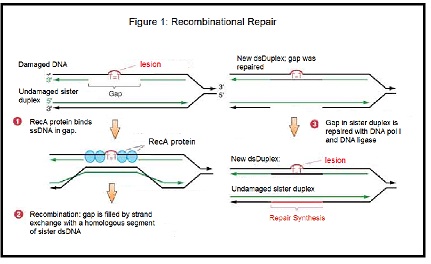
The recombinational repair pathway (Figure 1) and the RecA protein will be used to fill in this gap. The basic steps of the pathway are as follows:- The exposed single-stranded DNA is bound by RecA, forming a RecA-DNA filament.
- This single-stranded DNA-RecA filament finds a portion of homologous double-stranded DNA duplex. This DNA duplex is composed of two strands; one of these strands is exactly the same as the single-stranded DNA bound by RecA, and the other is exactly complementary. RecA is bound to the single stranded portion of DNA on the damaged duplex; this strand is missing its “pairing partner” and needs to be paired with a strand that is exactly complementary. The complementary portion that is added will fill in the gap on the damaged strand.
- RecA catalyzes the pairing of the single-stranded DNA to the one strand of the DNA duplex that is complementary. The other strand of the DNA duplex is displaced. This step is the actual "strand exchange."
Recombination is a very powerful event, and could cause problems if it happened in inappropriate situations. Recombination can only be allowed to occur between areas of homologous DNA, or the integrity of the DNA would be compromised. The occurrence of strand exchange relies heavily on the pairing of the RecA-bound single-stranded DNA with a strand of the duplex DNA; strand exchange can only occur if these strands are complementary and Watson-Crick base pairs are formed.
RecA function in the SOS Response
The SOS Response is triggered in a cell after DNA damage has occurred. If triggered, this response leads to the production of proteins that are needed for DNA repair. RecA is involved in the activation of this response; here RecA functions as a coprotease. This means that the binding of RecA to other proteins causes them to cleave themselves apart. In a healthy cell, LexA is bound to the cell's DNA. LexA is a repressor protein. This means that when LexA is bound to DNA, the transcription of genes involved in DNA repair is prevented. These genes are not transcribed or translated into protein. The transcription of these DNA repair genes is only activated when LexA cleaves itself, falls off the DNA and exposes the genes for transcription. The autocatalytic cleavage of LexA is possible only in the presence of a RecA-DNA filament that forms after DNA damage has occurred. In this way, RecA allows the expression of genes encoding the proteins necessary to repair DNA damage. This means these proteins are only translated if DNA damage has occurred.
Monomeric RecA
Monomeric RecA is comprised of 352 amino acids and has a molecular mass of 37,842 Daltons. The protein can be separated into three main domains; the amino terminus, the 240 amino acid core domain, and the carboxy domain. The large core domain contains amino acids important in RecA's ATPase function. RecA is a DNA-dependent ATPase, which means it can hydrolyze ATP when it is bound to DNA. The hydrolysis of ATP allows the RecA to dissociate from the RecA filament at the 5' end. Two areas of the core domain, the L1 and L2 loops, coordinate single-stranded DNA binding.
RecA as a Filament
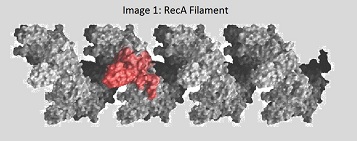
In its functional form, RecA acts as a filament (Image 1). Throughout the strand exchange reaction, there are three different types of RecA filaments.- When RecA first binds to ssDNA, it is called a “presynaptic filament.” This presynaptic filament finds the homologous DNA duplex
- Synaptic filament is the intermediate between the pre and postsynaptic filament. The single-stranded DNA is aligned with the DNA duplex and strand exchange begins.
- The postsynaptic filament is formed after the strand exchange reaction. RecA is still bound to the original strand from the presynaptic filament; the original strand is now bound to it's complementary strand from the donor duplex.
ATP and DNA in the RecA Filament
| |||||||||||