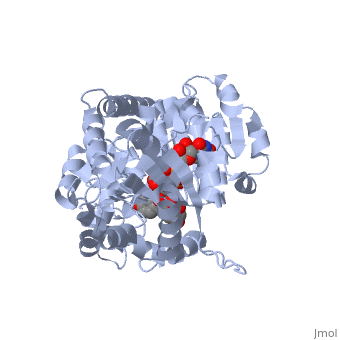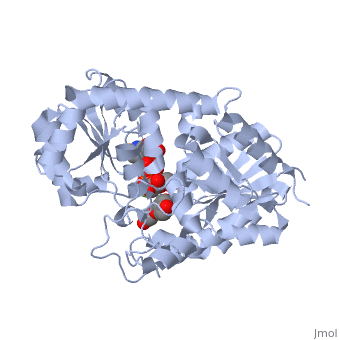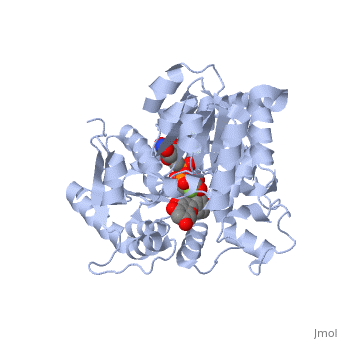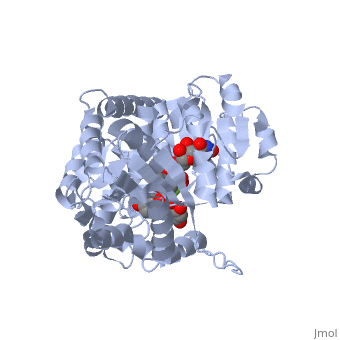
Vitis vinifera Flavonoid 3-O-Glucosyltransferase (Vv3GT) is involved in the modification of grape anthocyanins (a plant pigment) and thus could affect their water solubility and color stability [1]. The addition of a sugar molecule on the anthocyanin is a preliminary step to its transport to the cell vacuole. The transfer to the vacuole is important for the pigment accumulation. The anthocyanin accumulation plays a significant role in quality of agricultural produce, as it affects fruit color and its health benefits as a natural antioxidant. This enzyme affects the quality of both table grapes and wine grapes.
Introduction
Vv3GT belongs to Glycosyltransferases (GTs), a large family of enzymes involved in the transfer of sugar residues from a sugar donor to various substrates. Glycosylation of metabolites in plants is usually catalyzed by glycosyltransferases (GTs) belonging to the GT1 sub-family, as classified by the CAZy database , which use UDP-activated sugars as the major donor molecule and are thus referred to as UGTs. The Glycosyltransferase activity is highly important for the synthesis of thousands of plant metabolites. This enzymes differ in their specificity for substrate, position of glycosylation on the substrate and recognition of sugar donor. These differences are important and could affect the function and stability of the metabolite.
Relevance
A good example for the importance of glycosyltransferase specificity is shown in Citrus [2] . the difference in the position of glycosylation affects fruit flavor. Glycosylation of one position lids to the synthesis of bitter substances while the addition of same molecule at a different position leads to the synthesis of tasteless substances.
Structural highlights
Despite low primary sequence similarity, the secondary and tertiary structures of GTs are highly conserved [3].
GT-B
Plant glycosyltransferases assume one of two folds, GT-A or GT-B. Vv3GT is a GT-B enzyme[4]. GT-B enzymes consists of two β/α/β Rossmann-like domains [5]. The two domains are associated and face each other with the active-site lying between them. These domains are associated with the donor and acceptor substrate binding sites.
The , linked by a flexible loop and α-helix with a hinge region. The linker could be important for sugar donor binding. This linker region holds the UDP-sugar donor next to the C-terminal PSPG motif. The C-terminal domain is highly conserved and binds the UDP sugar donor (e. g. UDP-Glc). On the other hand, the N-terminal domain is not conserved it binds the substrate and provides catalytically active amino acids.
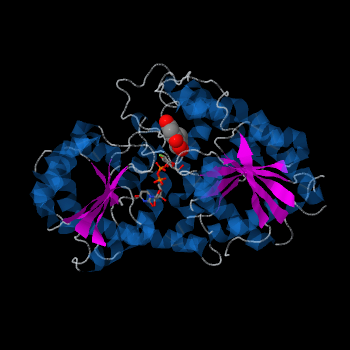
Vv3GT with two Rossmann-like domains
PSPG
The plant UGTs are characterized by sharing a highly conserved 44 amino acid motif referred to as the PSPG motif (Plant Secondary Product Glycosyltransferase motif). Amino acids of the PSPG motif provide most of the interactions with the sugar donor molecule. 10 highly conserved residues of the 44 amino acid PSPG motif are observed to directly interact with the UDP-sugar. In Vv3GT the motif begins with Pro 334 and ends with Gln 375.
Three conserved motifs involved in sugar binding are present in Vv3GT. The first, a motif (Thr 141; Ala 142) involved in sugar binding. The second, a (Trp 353; Asn 354; Ser 355) motif residues are involved in binding UDP phosphates. The third, motif residues also involved in sugar binding (Asp 374; Gln 375). The WNS and D/EQ motifs are part of the highly conserved PSPG region.
3D structure of glucosyltransferase
Glycosyltransferase
Quiz