Sandbox Reserved 1131
From Proteopedia
|
| This Sandbox is Reserved from 15/12/2015, through 15/06/2016 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1120 through Sandbox Reserved 1159. |
To get started:
More help: Help:Editing |
(Human) Phosphohistidine phosphatase 1 (PHPT1) belongs to the Janus family, it has 2 isoforms produced by alternative splicing, and 6 transcripts. It is encoded by the PHPT1 gene, located on the 9th chromosome.[1] This enzyme has been recently discovered so it has not been fully understood and characterized yet.
| Ligands: | |
| NonStd Res: | |
| Gene: | PHPT1, PHP14 (HUMAN) |
| Resources: | FirstGlance, OCA, PDBe, RCSB, PDBsum, TOPSAN |
Contents |
Structure
|
Generality
Phosphohistidine phosphatase 1 is a 14kDa homotrimeric protein, whose monomers contain 125 amino acids. Furthermore, one monomer contains 4 α helices, 6 β strands and 2 turns. [1]
Sequence of the PHPT1 protein
>>sp|Q9NRX4|PHP14_HUMAN 14 kDa phosphohistidine phosphatase OS=Homo sapiens GN=PHPT1 PE=1 SV=1 MAVADLALIPDVDIDSDGVFKYVLIRVHSAPRSGAPAAESKEIVRGYKWAEYHADIYDKV
SGDMQKQGCDCECLGGGRISHQSQDKKIHVYGYSMAYGPAQHAISTEKIKAKYPDYEVTW
ANDGY
Domains
PHPT1 contains an N-acetylation site on the residue Ala(2). There is also an Ocnus region (from His(6) to Asp(95)) which is characteristic of the Janus family. [2] It has one substrate binding site (see below), and one proton acceptor active site (His(53)).[1]
Function
It is an hydrolase. This characteristic structure allows it to have many activities : phosphoprotein and phosphohistidine phosphatase, calcium channel inhibition, ion channel binding. It is located in the cytosol and in extracellular exosome. [1]
Active site
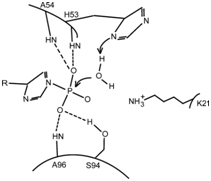
The active site is located between the beginning of helix α1 and loop L5. The catalytic residue is His(53), and the anchor sites of P(i)are the amine groups of His(53), Ala(54) and Ala(96) and the Hydroxy group of Ser(94), which form hydrogen bonds with it. [3]
Ligands and binding
There are 6 SO4 lingands, called SO4 201 to 206. 2 lingands bind to each monomer (2ai6), inducing a conformational change of all the monomers (2hw4). Then, the lingands bind to each other, forming a trimer (2NMM). SO4 is a 96 Da molecule, with two negative charges. The four oxygens allow good fixation on the monomers thanks to hydrogen bonds. The substrate is the P(i) of a histidine-phosphorylated residue, which will bind the active site. [4]
Catalyic mechanism
The P(i) substrate binds to the active site thanks to the formation of four H-bonds. The imidazole ring of His(53) acts as a base to activate a water molecule, which will in turn do a nucleophilic attack on the substrate. The amine group of the residue Lys(21) can help stabilize the transition state. [3]
Degradation of defective PHPT1
A possible degratation of PHPT1, thanks to quality control and cellular system self-guarding, can happen at two levels. A primary degradation of non-sens mutated PHPT1 mRNA is possible at the RNA level. Then, the formed protein can be destroyed by the ptoteasome if it is defective. [5]
Implication in biological functions and pathways
Histidine reversible phosphorylation plays important roles in several signal transductions and other cellular functions.
Tissue expression
It is usally expressed in the cytosol of cells located in the heart, in skeletal muscles, but also in the liver, in the pancreas and in the kidney. [2]
Inhibition of epithelial Ca²⁺ channel TRPV5
This channel is activated by the kinase NDPK-B, which phosphorylates his(711) in the C-terminal tail of TRPV5. PHPT1 inhibits the activity of TRPV5 by dephosphorylating the same residue, resulting in a decrease in Ca²⁺ flux. This mechanism plays a role in the regulation of Ca²⁺ reabsorption by the kidney. [6]
Inhibition of the K+ channel KCa3.1
PHPT1 dephosphorylates his(358) on KCa3.1, a Ca²⁺-dependant K+ channel that is activated by the phosphorylation on the same residue by NDPK-B. KCa3.1 helps maintain the negative membrane potential, and plays an important role in CD4 T cells signalling. This dephosphorylation inhibits the activity of the KCa3.1 channel and decreases the Ca²⁺ afflux, negatively regulating human CD4 T cells. [7]
Impact on the rate of dead cells
Several studies have shown the possible implication of Phosphohistidine phosphatase in cellular death. When the PHPT1 gene is knocked out, the rate of cellular death remains unchanged. However, an overexpression of the PHPT1 gene causes a decrease of the ATP-citrate lyase (ACL) activity due to the loss of a phosphate group on its His(760) which inactivates it. This inactivation increases the number of cellular deaths. This effect has been proved for neurons but not for others kind of cells. [8]
NB:dephosphorylation on a non-histidine residue
A study has proved that PHPT1 dephosphorylates histone H1 and polylysine, which do not contain Histidine residue, meaning that the protin has a broader specificity than the one that we know yet. [9]
Diseases and medical applications
Lung Cancer
It has been proved that PHPT1 concentration is linked to lung cancer. Indeed, PHPT1 is associated with the carcinogenesis and metastasis of this cancer, it promotes cell migration and invasion. In cancerous cells, the expression of PHPT1 is almost two fold the one in normal cells. Therefore, a therapy has been recently developped, aiming the inhibition or the silencing of PHPT1, and it is hoped to be benefic for lung cancer patients. [10]
Diabetes and regulation of the synthesis of insulin
Phosphohistidine phosphatase is expressed in the cells of the Langerhans’ islets (pancreas). The cytoplasm localization of Phosphohistidine phosphatase is not affected by the concentration of glucose in the cell. However, the expression of PHPT1 gene increases when the concentration of glucose is too high: Phosphohistidine phosphatase seems to be necessary for the synthesis of insulin (especially in the Mas-insulin secretion). If the Phosphohistidine phosphatase is non-functional or inhibited, all the mechanisms of the insulin synthesis are impacted: insulin is no longer produced and the rate of sugar in the blood is not regulated anymore. [11]
See also
References
- ↑ 1.0 1.1 1.2 1.3 http://www.uniprot.org/uniprot/Q9NRX4
- ↑ 2.0 2.1 http://www.genecards.org/cgi-bin/carddisp.pl?gene=PHPT1&keywords=PHPT1,Gene
- ↑ 3.0 3.1 Gong W, Li Y, Cui G, Hu J, Fang H, Jin C, Xia B. Solution structure and catalytic mechanism of human protein histidine phosphatase 1. Biochem J. 2009 Mar 1;418(2):337-44. PMID:18991813 doi:BJ20081571
- ↑ https://www.ebi.ac.uk/pdbe/entry/pdb/2nmm/biology
- ↑ Inturi R, Waneskog M, Vlachakis D, Ali Y, Ek P, Punga T, Bjerling P. A splice variant of the human phosphohistidine phosphatase 1 (PHPT1) is degraded by the proteasome. Int J Biochem Cell Biol. 2014 Dec;57:69-75. doi: 10.1016/j.biocel.2014.10.009., Epub 2014 Oct 14. PMID:25450458 doi:http://dx.doi.org/10.1016/j.biocel.2014.10.009
- ↑ Cai X, Srivastava S, Surindran S, Li Z, Skolnik EY. Regulation of the epithelial Ca(2)(+) channel TRPV5 by reversible histidine phosphorylation mediated by NDPK-B and PHPT1. Mol Biol Cell. 2014 Apr;25(8):1244-50. doi: 10.1091/mbc.E13-04-0180. Epub 2014, Feb 12. PMID:24523290 doi:http://dx.doi.org/10.1091/mbc.E13-04-0180
- ↑ Srivastava S, Zhdanova O, Di L, Li Z, Albaqumi M, Wulff H, Skolnik EY. Protein histidine phosphatase 1 negatively regulates CD4 T cells by inhibiting the K+ channel KCa3.1. Proc Natl Acad Sci U S A. 2008 Sep 23;105(38):14442-6. doi:, 10.1073/pnas.0803678105. Epub 2008 Sep 16. PMID:18796614 doi:http://dx.doi.org/10.1073/pnas.0803678105
- ↑ Klumpp S, Faber D, Fischer D, Litterscheid S, Krieglstein J. Role of protein histidine phosphatase for viability of neuronal cells. Brain Res. 2009 Apr 6;1264:7-12. doi: 10.1016/j.brainres.2008.12.052. Epub 2008, Dec 30. PMID:19138678 doi:http://dx.doi.org/10.1016/j.brainres.2008.12.052
- ↑ Ek P, Ek B, Zetterqvist O. Phosphohistidine phosphatase 1 (PHPT1) also dephosphorylates phospholysine of chemically phosphorylated histone H1 and polylysine. Ups J Med Sci. 2015 Mar;120(1):20-7. doi: 10.3109/03009734.2014.996720. Epub 2015, Jan 9. PMID:25574816 doi:http://dx.doi.org/10.3109/03009734.2014.996720
- ↑ Xu AJ, Xia XH, DU ST, Gu JC. Clinical significance of PHPT1 protein expression in lung cancer. Chin Med J (Engl). 2010 Nov;123(22):3247-51. PMID:21163124
- ↑ Kamath V, Kyathanahalli CN, Jayaram B, Syed I, Olson LK, Ludwig K, Klumpp S, Krieglstein J, Kowluru A. Regulation of glucose- and mitochondrial fuel-induced insulin secretion by a cytosolic protein histidine phosphatase in pancreatic beta-cells. Am J Physiol Endocrinol Metab. 2010 Aug;299(2):E276-86. doi:, 10.1152/ajpendo.00091.2010. Epub 2010 May 25. PMID:20501872 doi:http://dx.doi.org/10.1152/ajpendo.00091.2010