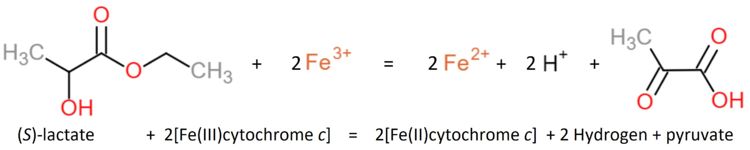The oxidoreductase Flavocytochrome b(2) (Arg289Lys mutant), categorized as EC: 1.1.2.3 according to IUB, is encoded in the Saccharomyces cerevisiae (strain ATCC 204508 / S288c) gene CYB2.
Function
Flavocytochrome b(2) (Arg289Lys mutant) from Saccharomyces cerevisiae is an oxidoreductase that couples dehydrogenation of L-lactate to cytochrome c reduction by electron transfer. It is one reaction of the bacterial lactate metabolic pathway localized in the mitochondrial intermembrane space.
Reaction
Flavocytochrome b(2) catalyzes dehydrogenation of L-lactate and the coupled cytochrome c reduction.
Similar oxidants that can be used to perform the reaction in vitro are ferricyanide, phenazine methosulfate and quinone (experiment first performed 1963 by Nygaard[1], later in 1966 described by Symons and Burgoyne[2]).
The yeasts L-lactate dehydrogenase can be inhibited by heavy metals, oxygen, glycerate, oxalate, malate, phenylpyruvate and fatty acids (Nygaard, 1963[1]).
The enzyme shows a specificity for L-lactate but none for the D-isomer or α-hydroxybutyrate.
Structure highlight
Flavocytochrome b(2) is a tetrameric enzyme (Jacq and Lederer, 1972 and 1974[3],[4]). Each of the four identical subunits is composed by one single polypeptide chain.
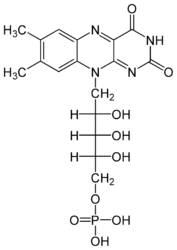
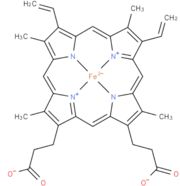
Each subunit contains a binding site for the selectively non-covalently binding of the cofactor FMN(3-) (Flavinmononucleotide), as well as one in with the iron complexed in the tetrapyrrole ring interacts with heme b(2-) cofactor (Risler and Groudinsky, 1973[5]).
The amino acid sequence in the heme binding region was first determined by Guidard et al, 1974[6].
For every subunit of the wild type protein form, the crystallized preparation analysis determined a molecular weight of the α-chain of 36 kD (Appleby and Morton, 1959[7]) and the β-chain of 21 kD (Jacq and Lederer, 1974[8]).
The sulfite adduct recombinant enzyme produced when expressed in E. coli was also crystallized (Tegoni and Cambillau, 1994[9]) so key active site residues could be identified and comparisons with the mutant protein can be made.
The Arg289 of the E. coli wild type sulfite adduct can adopt two different conformations, in one of which its side chain is stacked against Arg376, that directly interacts with the substrate, while in the second one the Arg289 side chain points towards the active site.
The mutation changes that Arg289 into a Lysine becoming R289K-b(2). The mutant ARG289LYS can still be found in both conformations but it is now changing the kinetics of the reactions (Tegoni and Cambillau, 1994[9]).
It is rising the Ki of several components in comparison to the wild type, while kcat and KM are also changed by a factor of 10.
It changes also the induction by L-lactate. Patterns of inhibition by pyruvate and oxalate are altered and the enzyme stops being inhibited by substrate excess.
The mutation has altered the flavin reduction, the first step of the catalytic cycle. It is shown that the mutation enhances the stability of both enzyme-substrate-complex and the transition state, as well as in ligand binding to the active site when the flavin is in the semiquinone state. The first electron transfer step, from the reduced flavin to heme, is not affected by the mutation, but it indeed affects the second electron transfer from flavin semiquinone to heme b(2).
The resolution through X-ray crystal structure R289K-b(2) has been determined to 2,75 Å.