2emt
From Proteopedia
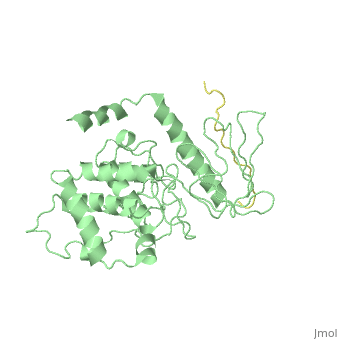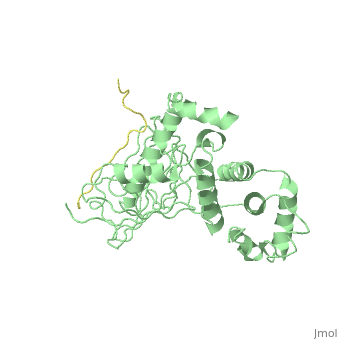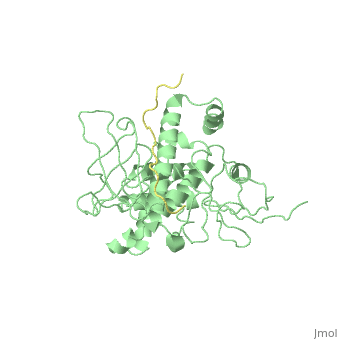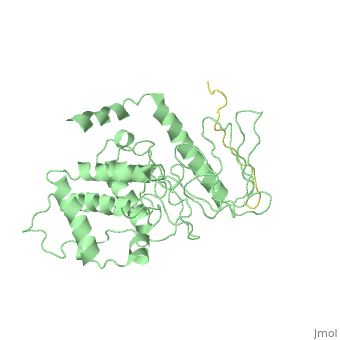
Crystal Structure Analysis of the radixin FERM domain complexed with adhesion molecule PSGL-1
Structural highlights
FunctionRADI_MOUSE Probably plays a crucial role in the binding of the barbed end of actin filaments to the plasma membrane. Evolutionary ConservationCheck, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedP-selectin glycoprotein ligand-1 (PSGL-1), an adhesion molecule with O-glycosylated extracellular sialomucins, is involved in leukocyte inflammatory responses. On activation, ezrin-radixin-moesin (ERM) proteins mediate the redistribution of PSGL-1 on polarized cell surfaces to facilitate binding to target molecules. ERM proteins recognize a short binding motif, Motif-1, conserved in cytoplasmic tails of adhesion molecules, whereas PSGL-1 lacks Motif-1 residues important for binding to ERM proteins. The crystal structure of the complex between the radixin FERM domain and a PSGL-1 juxtamembrane peptide reveals that the peptide binds the groove of FERM subdomain C by forming a beta-strand associated with strand beta5C, followed by a loop flipped out towards the solvent. The Motif-1 3(10) helix present in the FERM-ICAM-2 complex is absent in PSGL-1 given the absence of a critical Motif-1 alanine residue, and PSGL-1 reduces its contact area with subdomain C. Non-conserved positions are occupied by large residues Met9 and His8, which stabilize peptide conformation and enhance groove binding. Non-conserved residues play an important role in compensating for loss of binding energy resulting from the absence of conserved residues important for binding. Structural basis of PSGL-1 binding to ERM proteins.,Takai Y, Kitano K, Terawaki S, Maesaki R, Hakoshima T Genes Cells. 2007 Dec;12(12):1329-38. PMID:18076570[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
| ||||||||||||||||||