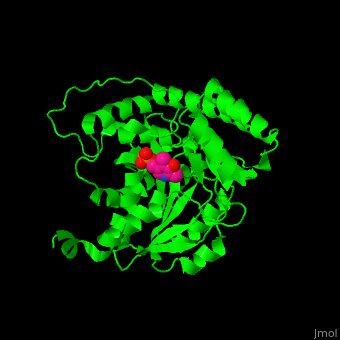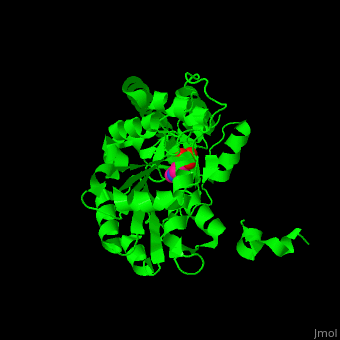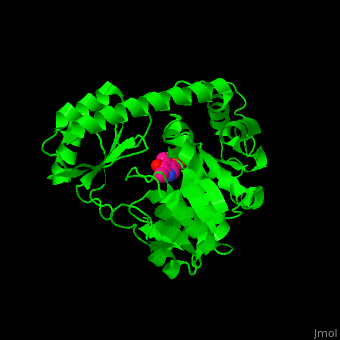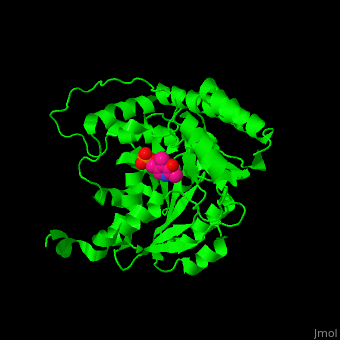
Aminotransferase
From Proteopedia
| |||||||||||
References
- ↑ Mizuguchi H, Hayashi H, Miyahara I, Hirotsu K, Kagamiyama H. Characterization of histidinol phosphate aminotransferase from Escherichia coli. Biochim Biophys Acta. 2003 Apr 11;1647(1-2):321-4. PMID:12686152
- ↑ Kirsch JF, Eichele G, Ford GC, Vincent MG, Jansonius JN, Gehring H, Christen P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J Mol Biol. 1984 Apr 15;174(3):497-525. PMID:6143829 doi:http://dx.doi.org/10.1016/0022-2836(84)90333-4
- ↑ Kirsch JF, Eichele G, Ford GC, Vincent MG, Jansonius JN, Gehring H, Christen P. Mechanism of action of aspartate aminotransferase proposed on the basis of its spatial structure. J Mol Biol. 1984 Apr 15;174(3):497-525. PMID:6143829 doi:http://dx.doi.org/10.1016/0022-2836(84)90333-4
- ↑ Lee M, Sousa MC. Structural Basis for Substrate Specificity in ArnB. A Key Enzyme in the Polymyxin Resistance Pathway of Gram-Negative Bacteria. Biochemistry. 2014 Feb 4;53(4):796-805. doi: 10.1021/bi4015677. Epub 2014 Jan 24. PMID:24460375 doi:http://dx.doi.org/10.1021/bi4015677
- ↑ Schoenhofen IC, Lunin VV, Julien JP, Li Y, Ajamian E, Matte A, Cygler M, Brisson JR, Aubry A, Logan SM, Bhatia S, Wakarchuk WW, Young NM. Structural and functional characterization of PseC, an aminotransferase involved in the biosynthesis of pseudaminic acid, an essential flagellar modification in Helicobacter pylori. J Biol Chem. 2006 Mar 31;281(13):8907-16. Epub 2006 Jan 18. PMID:16421095 doi:10.1074/jbc.M512987200
- ↑ Larkin A, Olivier NB, Imperiali B. Structural Analysis of WbpE from Pseudomonas aeruginosa PAO1: A Nucleotide Sugar Aminotransferase Involved in O-Antigen Assembly . Biochemistry. 2010 Jul 28. PMID:20604544 doi:10.1021/bi100805b
- ↑ Haruyama K, Nakai T, Miyahara I, Hirotsu K, Mizuguchi H, Hayashi H, Kagamiyama H. Structures of Escherichia coli histidinol-phosphate aminotransferase and its complexes with histidinol-phosphate and N-(5'-phosphopyridoxyl)-L-glutamate: double substrate recognition of the enzyme. Biochemistry. 2001 Apr 17;40(15):4633-44. PMID:11294630
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Joel L. Sussman, Jaime Prilusky