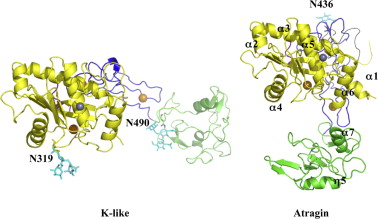
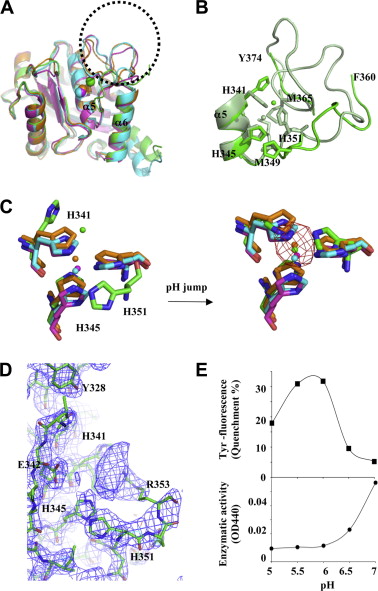
Atragin
From Proteopedia
| |||||||||||
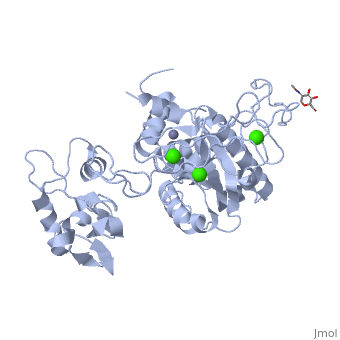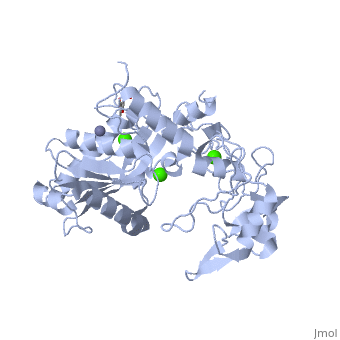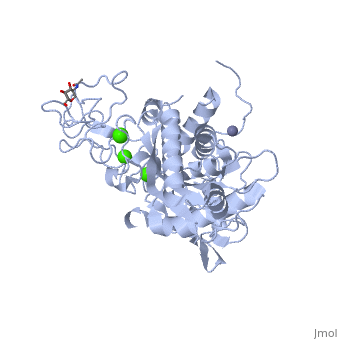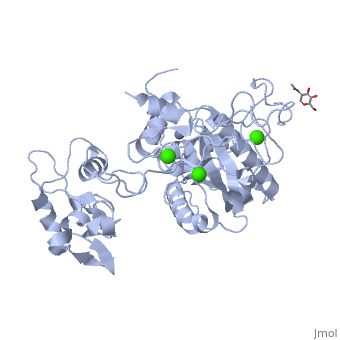
3D structures of atragin
3k7l, 3k7n - Atragin - chinese cobra
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Guan HH, Goh KS, Davamani F, Wu PL, Huang YW, Jeyakanthan J, Wu WG, Chen CJ. Structures of two elapid snake venom metalloproteases with distinct activities highlight the disulfide patterns in the D domain of ADAMalysin family proteins. J Struct Biol. 2010 Mar;169(3):294-303. Epub 2009 Nov 22. PMID:19932752 doi:10.1016/j.jsb.2009.11.009
- ↑ Fox JW, Serrano SM. Structural considerations of the snake venom metalloproteinases, key members of the M12 reprolysin family of metalloproteinases. Toxicon. 2005 Jun 15;45(8):969-85. Epub 2005 Apr 9. PMID:15922769 doi:10.1016/j.toxicon.2005.02.012
- ↑ Gutierrez JM, Rucavado A. Snake venom metalloproteinases: their role in the pathogenesis of local tissue damage. Biochimie. 2000 Sep-Oct;82(9-10):841-50. PMID:11086214
- ↑ 4.0 4.1 4.2 Igarashi T, Araki S, Mori H, Takeda S. Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins. FEBS Lett. 2007 May 29;581(13):2416-22. Epub 2007 Apr 30. PMID:17485084 doi:10.1016/j.febslet.2007.04.057
- ↑ Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003 Jan 1;17(1):7-30. PMID:12514095 doi:10.1101/gad.1039703
- ↑ White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003 Oct;15(5):598-606. PMID:14519395
- ↑ Moss ML, Jin SL, Milla ME, Bickett DM, Burkhart W, Carter HL, Chen WJ, Clay WC, Didsbury JR, Hassler D, Hoffman CR, Kost TA, Lambert MH, Leesnitzer MA, McCauley P, McGeehan G, Mitchell J, Moyer M, Pahel G, Rocque W, Overton LK, Schoenen F, Seaton T, Su JL, Becherer JD, et al.. Cloning of a disintegrin metalloproteinase that processes precursor tumour-necrosis factor-alpha. Nature. 1997 Feb 20;385(6618):733-6. PMID:9034191 doi:10.1038/385733a0
- ↑ Iwamoto R, Yamazaki S, Asakura M, Takashima S, Hasuwa H, Miyado K, Adachi S, Kitakaze M, Hashimoto K, Raab G, Nanba D, Higashiyama S, Hori M, Klagsbrun M, Mekada E. Heparin-binding EGF-like growth factor and ErbB signaling is essential for heart function. Proc Natl Acad Sci U S A. 2003 Mar 18;100(6):3221-6. Epub 2003 Mar 5. PMID:12621152 doi:10.1073/pnas.0537588100
- ↑ Asakura M, Kitakaze M, Takashima S, Liao Y, Ishikura F, Yoshinaka T, Ohmoto H, Node K, Yoshino K, Ishiguro H, Asanuma H, Sanada S, Matsumura Y, Takeda H, Beppu S, Tada M, Hori M, Higashiyama S. Cardiac hypertrophy is inhibited by antagonism of ADAM12 processing of HB-EGF: metalloproteinase inhibitors as a new therapy. Nat Med. 2002 Jan;8(1):35-40. PMID:11786904 doi:10.1038/nm0102-35
- ↑ Haga S, Yamamoto N, Nakai-Murakami C, Osawa Y, Tokunaga K, Sata T, Yamamoto N, Sasazuki T, Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc Natl Acad Sci U S A. 2008 Jun 3;105(22):7809-14. Epub 2008 May 19. PMID:18490652 doi:10.1073/pnas.0711241105
- ↑ Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, Torrey D, Pandit S, McKenny J, Braunschweiger K, Walsh A, Liu Z, Hayward B, Folz C, Manning SP, Bawa A, Saracino L, Thackston M, Benchekroun Y, Capparell N, Wang M, Adair R, Feng Y, Dubois J, FitzGerald MG, Huang H, Gibson R, Allen KM, Pedan A, Danzig MR, Umland SP, Egan RW, Cuss FM, Rorke S, Clough JB, Holloway JW, Holgate ST, Keith TP. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002 Jul 25;418(6896):426-30. Epub 2002 Jul 10. PMID:12110844 doi:10.1038/nature00878
- ↑ Wu E, Croucher PI, McKie N. Expression of members of the novel membrane linked metalloproteinase family ADAM in cells derived from a range of haematological malignancies. Biochem Biophys Res Commun. 1997 Jun 18;235(2):437-42. PMID:9199213 doi:10.1006/bbrc.1997.6714
- ↑ Muniz JR, Ambrosio AL, Selistre-de-Araujo HS, Cominetti MR, Moura-da-Silva AM, Oliva G, Garratt RC, Souza DH. The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: insights for a new classification of snake venom metalloprotease subgroups. Toxicon. 2008 Dec 1;52(7):807-16. Epub 2008 Sep 27. PMID:18831982 doi:10.1016/j.toxicon.2008.08.021
- ↑ 14.0 14.1 14.2 14.3 14.4 Takeda S, Igarashi T, Mori H, Araki S. Crystal structures of VAP1 reveal ADAMs' MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006 Jun 7;25(11):2388-96. Epub 2006 May 11. PMID:16688218
- ↑ Zhu Z, Gao Y, Zhu Z, Yu Y, Zhang X, Zang J, Teng M, Niu L. Structural basis of the autolysis of AaHIV suggests a novel target recognizing model for ADAM/reprolysin family proteins. Biochem Biophys Res Commun. 2009 Aug 14;386(1):159-64. Epub 2009 Jun 6. PMID:19505434 doi:10.1016/j.bbrc.2009.06.004
- ↑ Gomis-Ruth FX. Structural aspects of the metzincin clan of metalloendopeptidases. Mol Biotechnol. 2003 Jun;24(2):157-202. PMID:12746556 doi:10.1385/MB:24:2:157
- ↑ Odell GV, Ferry PC, Vick LM, Fenton AW, Decker LS, Cowell RL, Ownby CL, Gutierrez JM. Citrate inhibition of snake venom proteases. Toxicon. 1998 Dec;36(12):1801-6. PMID:9839664
- ↑ Marques-Porto R, Lebrun I, Pimenta DC. Self-proteolysis regulation in the Bothrops jararaca venom: the metallopeptidases and their intrinsic peptidic inhibitor. Comp Biochem Physiol C Toxicol Pharmacol. 2008 May;147(4):424-33. Epub, 2008 Feb 5. PMID:18325841 doi:10.1016/j.cbpc.2008.01.011