BASIL2023GVQ8DN35
From Proteopedia
Q8DN35 Protein
The predicted function of the Q8DN35 protein was studied during the spring 2023 semester at Grand View University by Lucas Kramer and Anie Rehbein in CHEM 453 - Biochemical Techniques.
|
Contents |
Computational Structure
Introduction
Q8DN35 is an enzyme with a computational structure but no known function. Our enzyme was predicted to be a kinase. The hypothesized function of this protein was that of a glucokinase, and through in silico research, two substrates were selected for further testing. Fructose-6-Phosphate (F6P) and N-acetyl-D-glucosamine (NAG) were selected based on binding affinity during PyRx docking analysis and based on correct binding site location. NetGO machine learning algorithms predicted the function of Q8DN35 to be a glucokinase, with N-acetylglucokinase activity also predicted deeply. Kinetic activity was analyzed to determine whether F6P or NAG were correct substrates. Bradford standards and gel electrophoresis were also conducted to determine concentration of Q8DN35 and to ensure the protein was present.
Function
Through computational docking results, the predicted function for the Q8DN35 protein is that of a glucokinase. The use of tools such as NetGO, BLAST, DALI, InterPRO, PyRX and SPRITE allowed for this predicted function. Q8DN35's sequence is related to that of the ROK Kinase family. NetGO predicted that this protein will have glucokinase activity as it's deepest result. PyRX docking data showed us that NAG (N-acetyl-D-glucosamine) and F6P (fructose-6-phosphate) had high binding affinity in the potential binding pocket of Q8DN35.
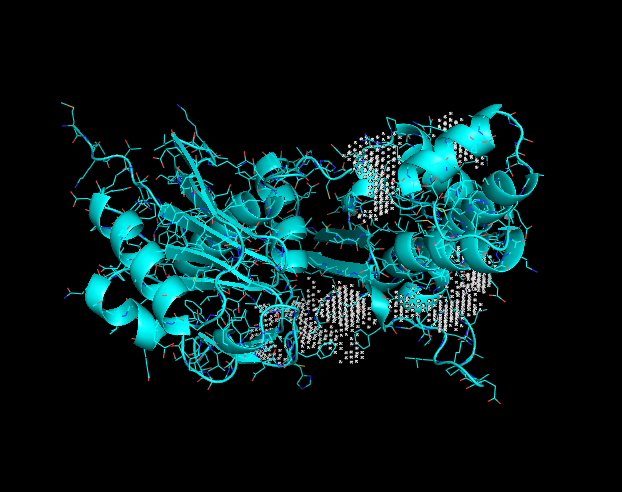
Binding site
This image displays potential binding sites to the Q8DN35 protein. Each grey "X" indicates a potential binding site. These binding sites were taken into consideration when looking at PyRx docking tools.
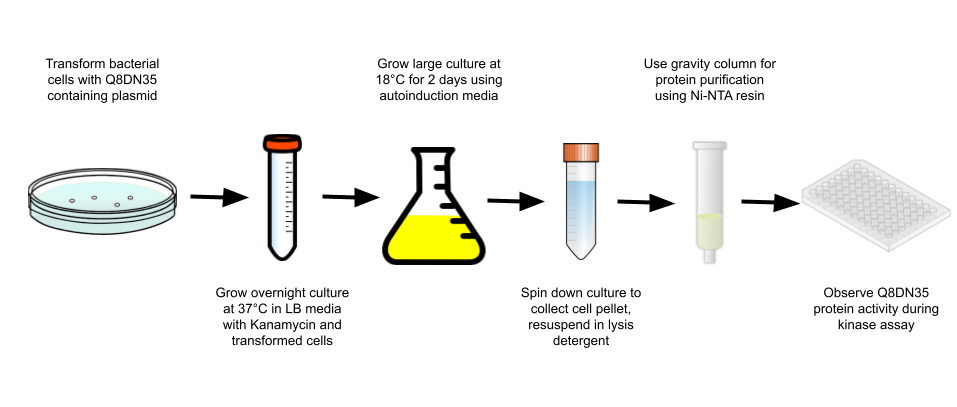
Experimental process
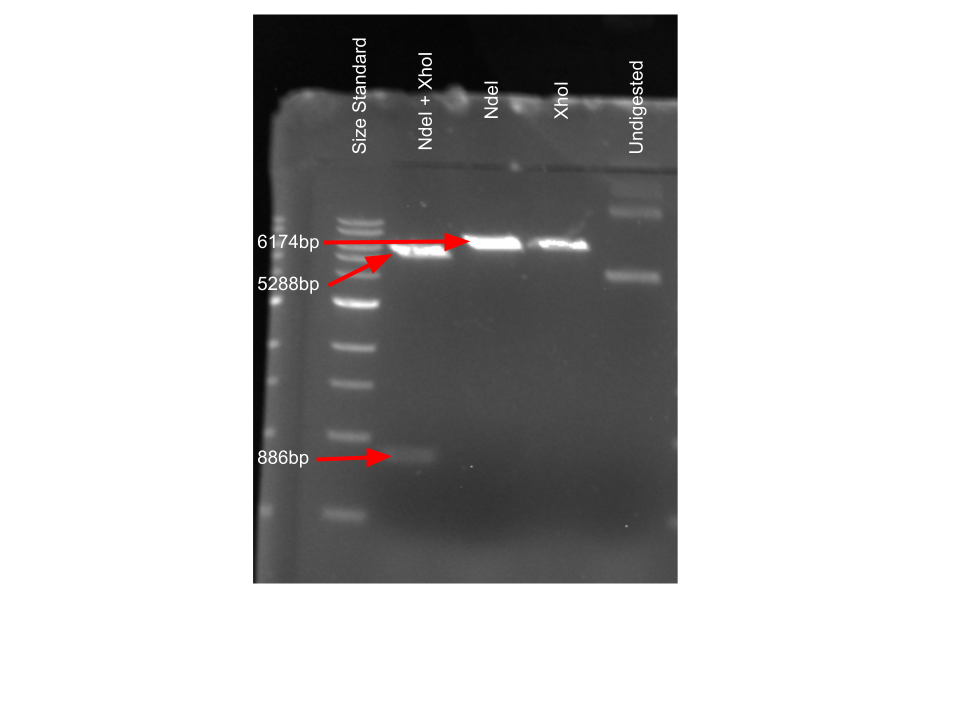
Q8DN35 DNA analysis
To confirm presence of Q8DN35 DNA, plasmid digestion was performed to observe correct DNA. Sizes were determined from looking at the plasmid map provided for our kinase enzyme DNA. Cutting with both of the chosen restriction enzymes yielded a smaller band containing our insert and a larger band containing the rest of the plasmid. Using only one restriction enzyme gives one band with all of the plasmid DNA present.
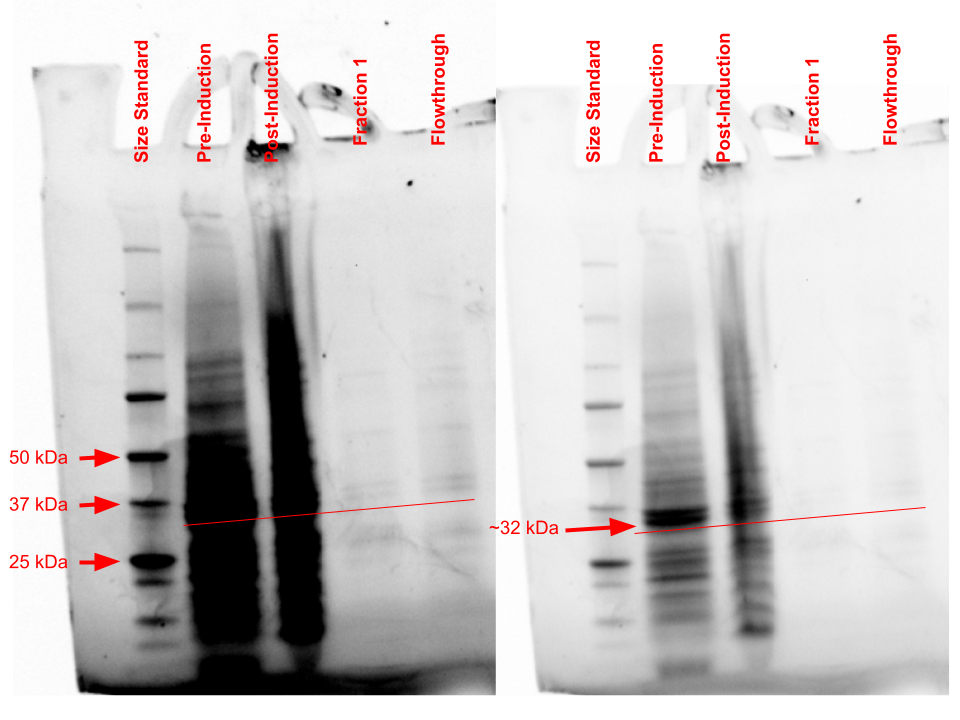
Protein purification analysis
Through the use of a gravity purification column and Ni-NTA resin, Q8DN35 was purified and a Bradford analysis was conducted to determine the amount of protein in the purified sample. This purification resulted in 0.055 mg/mL of protein being purified. Through SDS-PAGE analysis, observation of purified protein was hard to see in the fraction column but large banding occurred in the pre- and post-induction samples around 31.88 kDa, which is the predicted molecular weight for Q8DN35. Further analysis with more concentrated protein sample will need to be done to confirm correct protein positioning.
Kinase activity analysis
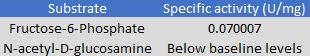
To observe Q8DN35's activity, a kinase activity was ran with 2 substrates to observe potential substrate usage. The 2 substrates that were tested are NAG and F6P. After kinetics calculations were finished, we conclude that NAG has a negative effect on Q8DN35 activity and F6P has a positive effect. The magnitude of increase in activity was not ideal, so further exploration into substrates and kinetic assay protocols will need to be done to determine the correct substrate for Q8DN35.
References
1.Miyazono, K., Tabei, N., Morita, S., Ohnishi, Y., Horinouchi, S., & Tanokura, M. (2012). Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus. Journal of bacteriology, 194(3), 607–616. https://doi.org/10.1128/JB.06173-11 2. The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.
Proteopedia Page Contributors and Editors (what is this?)
Lucas Kramer, Anie Rehbein, Jaime Prilusky, Bonnie Hall
DOI: https://dx.doi.org/10.14576/957644.4038169 (?)Citation: Kramer L, Rehbein A, Hall B, 2024, "BASIL2023GVQ8DN35",