From Proteopedia
proteopedia linkproteopedia link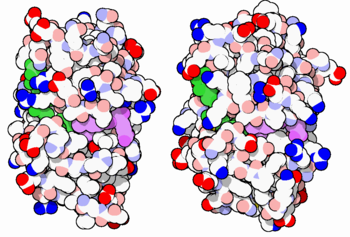
E. coli (left) and human (right) DHFR have a similar architecture and mode of binding to NADPH(green) and the competitive inhibitor methotrexate(purple). Original image by David Goodsell
The enzyme dihydrofolate reductase (DHFR) occurs in all organisms and has been particularly well-studied in the bacterium Escherichia coli and in humans[1][2][3]. It catalyzes the reduction of dihydrofolate to tetrahydrofolate, with NADPH acting as hydride donor. The human enzyme is a target for developing inhibitors used in anti-cancer chemotherapies[4], while the bacterial enzymes are targets for developing inhibitors as antibiotics. DHFR is a model enzyme for studying the kinetics, mechanism, and inhibition of enzymatic reactions and the underlying structure and conformational dynamics.
DHFR occurs in all organisms and most cells
DHFR is found in all organisms. Some bacteria acquire resistance to DHFR inhibitors through expressing a second form of DHFR coded on a plasmid. The enzymes from E. coli (ecDHFR) and humans (hDHFR) have similar folds, while the plasmid-encoded enzyme has an unrelated fold. In humans, DHFR is expressed in most tissues[1], and there are two genes, DHFR and DHFR2/DHFRL1, the latter targeted to mitochondria[5]. Mice and rats lack the second gene but also show DHFR activity in mitochondria[6].
Reactions catalyzed
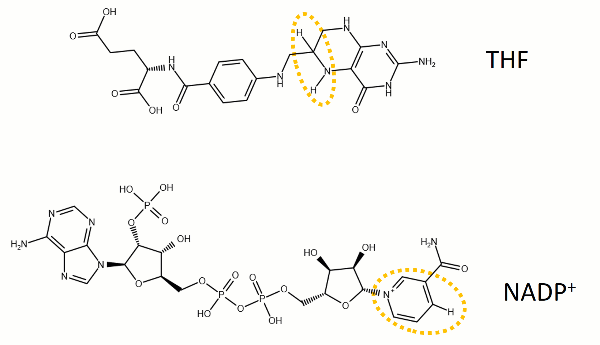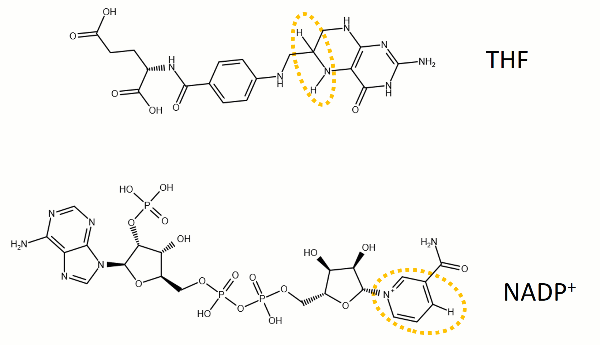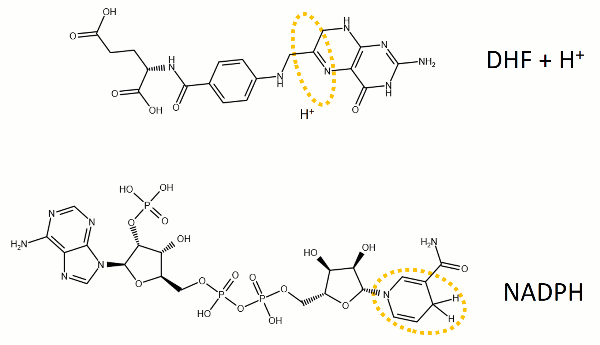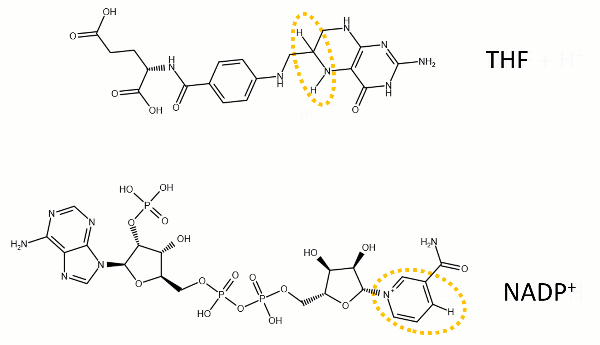
The reaction catalyzed by DHFR reduces a double bond in dihydrofolate (DHF) to form tetrahydrofolate (THF) by transfering a hydride from nicotinamide adenine dinucleotide phosphate (NADPH)
Dihydrofolate reductase (DHFR, 1.5.1.3 [2]) is an enzyme which uses the co-factor NADPH as electron donor. It catalyzes the reduction of dihydrofolic acid (DHF) to tetrahydrofolic acid (THF) as NADPH is oxidized to NADP+. The mammalian enzymes also accept folic acid as a substrate, reducing it to THF. This allows the use of folic acid, which is easier to synthesize than DHF or THF, to fortify food.[7][8]. Some bacterial enzymes also accept folic acid as a substrate [9] but it acts as a competitive inhibitor in the E. coli enzyme.
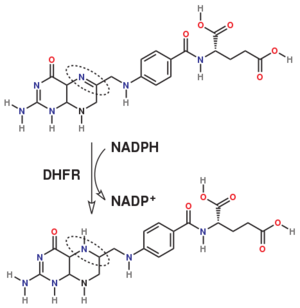
The folate is a form of the essential vitamin B9. Folate is not part of our natural diet (it contains dihydrofolate and tetrahydrofolate, sometimes as a poly-glutamate conjugate) but is bioavailable and simpler to synthesize.
Relevance
Tetrahydrofolate (THF) is an essential cofactor of
one-carbon metabolism[10][11].
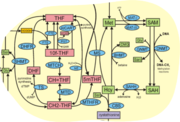
Metabolites and enzymes of one-carbon metabolism. Metabolites shown in red (DHF, 10f-THF, CH=THF, CH2-THF, 5mTHF) are all related to THF (by oxidation or methylation)
For example, THF is required for turning homocysteine into the amino acid methionine, and for biosynthesis of dTTP, one of the four nucleotide building blocks of DNA, from dUTP. In these reactions, tetrahydrofolate is first methylated and then oxidized to dihydrofolate. To allow for multiple rounds of turnover, dihydrofolate has to be reduced again; dihydrofolate reductase is the enzyme that enables this. Like many common cofactors, tetrahydrofolate is not synthesized de novo in the human body. Instead, it is provided as vitamin B9 in a healthy diet, for instance through leafy vegetables (the name for folic acid come from e.g. Latin folio, leaf). Many countries fortify common food ingredients like flour with vitamin B9 (in the form of folic acid) to ensure sufficient dietary levels. As described above, DHFR also plays a role in reducing folic acid to the biologically active tetrahydrofolate.
Structure and Function
| Fold and ligand binding
DHFR from bacteria and vertebrates share a common fold (CATH, SCOP) featuring a surrounded by . The bind in similar locations in bacterial and vertebrate enzymes. The substrate (and the competive inhibitors ) bind on one side of the beta sheet while the co-factor NADPH wraps around the sheet (around the C-terminal ends of strands as found in the classic Rossmann fold nucleotide binding site), with the nicotinamide moiety next to the substrate and the adenine moiety on the other side of the sheet.
In some cases, DHFR forms a complex with the enzymes that produce dihydrofolate, e.g. thymidylate synthase (TS)[12].
Mechanism
DHFR is thought to proceed in a multi-step mechanism. Once NADPH and DHF are bound to the E. coli enzyme, DHF is first protonated and then reduced through a hydride transfer from NADPH.[13] Substrate binding and product release is thought to have a definite choreography, with fresh NADPH binding before THF is released.
The changes during the catalytic cycle. The enzyme has an occluded conformation where a loop blocks entry of NADPH into the active site, and a closed conformation, where NADPH and DHF are in close contact.
A model by M. R. Sawaya and J. Kraut from 1997 based on six isomorphous crystal structures shows the envisioned sequence of events in the movie below [14]. The original web page of this movie is available on the web archive.
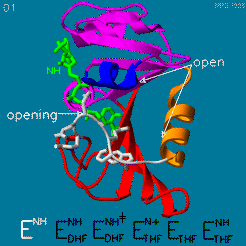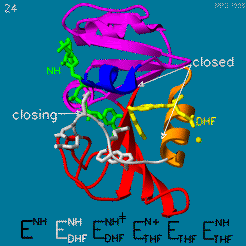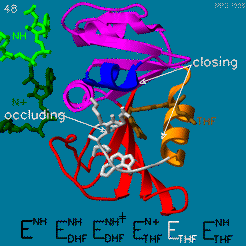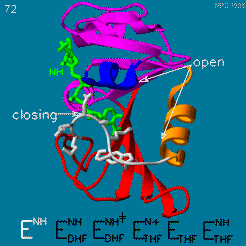
The was studied using neutron diffraction and high-resolution X-ray diffraction to visualize the hydrogen atoms that are usually not visible in crystal structures.[15]
The hydride transfer is thought to involve hydride tunneling, supported by temperature-dependent kinetic isotope effects[16]. Tunneling is a quantum phenomenon explaining how a small particle can cross an activation barrier even when it lacks sufficient activation energy. [17]
Inhibitors
The human enzyme is sufficiently different from the bacterial enzymes to make specific inhibitors[18]. Inhibitors of the human enzyme are used in cancer treatment while inhibitors of the bacterials enzymes[19] are used as broad-spectrum antibiotics or more specifically to treat a specific infectious disease. An example of in inhibitor of bacterial DHFR is Trimethoprim (TMP).
Antifolate inhibitors like (MTX), which is very similar to , are used in cancer therapy, see also Methotrexate.
Resistance
Antibiotics that inhibit bacterial DHFR are widely used in medicine and agriculture, and some bacterial strains have acquired resistance[20]. For example, some bacteria carry (either on their genome or on a plasmid) the trimethoprim-resistant dfrB gene coding for an enzyme DfrB that has a fold distinct from DHFR (coded by the folA gene)[21]. Like DHFR, Dfrb has dihydrofolate reductase activity, but unlike DHFR, it is not inhibited by trimethoprim. Thus, resistant bacteria can grow even in the presence of Trimethoprim, relying on the Dfrb enzyme while the DHFR enzyme is inhibited. The is unique in that the active site has fourfold symmetry, with DHF and NADPH bound by the same amino acid residues from different subunits. In the available crystal structure, two folate molecules are bound (use checkbox to show). The enzyme is non-specific and can bind to unproductive combinations of ligands (two DHF molecules or two NADPH molecules) or to the productive combination of one DHF and one NADPH molecule each.
|
See also
3D Structures of Dihydrofolate reductase
Dihydrofolate reductase 3D structures
Acknowledgements
This page was revised as part of the Spring 2022 Biochemistry II course at WSU. Thanks go to Kia, Anna, Shaylie and Michael for helpful suggestions to improve the page.
References





