Function
GTPase KRas (KRas) plays a critical role in the control of cellular growth. Kras stands for Kirsten rat sarcoma virus gene. KRas is a small GTPase that functions as molecular switches by alternating
between inactive GDP-bound state and active GTP-bound state[1].
KRas4A and KRas4B are two isoforms of KRas differing in 15 residues which are involved in trafficking and in membrane localisation[2].
Disease
A single mutation of KRas G12C or KRas G12V activates KRas and is implicated with various malignancies including lung adenocarcinoma, mutinous adenoma, ductal carcinoma of the pancreas and colorectal carcinoma[3]. KRas mutations are the most common genetic abnormalities in cancer.
Relevance
KRas inhibition represents an attractive therapeutic strategy for many cancers.
Structural highlights
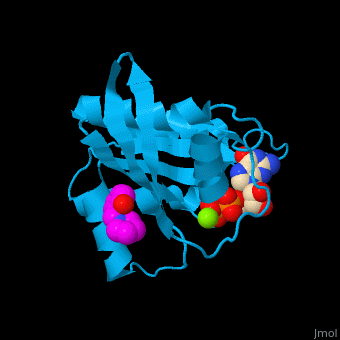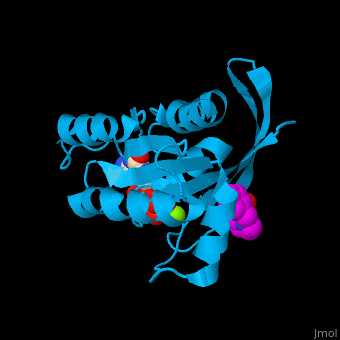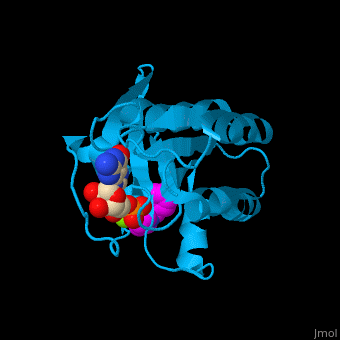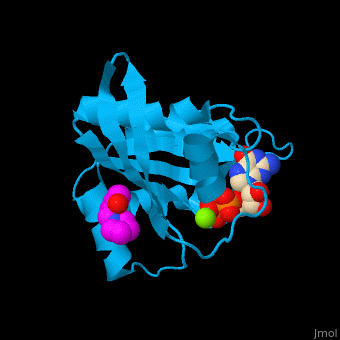
The GDP-bound KRas binds a following conformational change of the protein[4]. Water molecules are shown as red spheres.
3D Structures of GTPase KRas
GTPase KRas 3D structures