Isochorismate pyruvate lyase
From Proteopedia
Contents |
Isochorismate Pyruvate Lyase
Gateway for Substrate Entry and Product Exit
Isochorismate
Isochorismate is the end product of the shikimate pathway that is essential for the synthesis of many primary and secondary metabolites. It is synthesized from chorismate by isochorismate synthase enzyme. Chorismate is the common precursor for synthesis of aromatic amino acids, cofactors, phenazines and siderophores. Many enzymes are involved in the pathway and these enzymes are present in microbes and plants and absent in mammals and provide potential targets for antimicrobial drugs and herbicides.Isochorismate pyruvate Lyase (IPL) is one such enzyme[1].
Function
It is involved in the catalysis of the transformation of isochorismate to pyruvate and salicylate. This reaction is the committed step in the biosynthesis of salicylate-based siderophores in several pathogenic bacteria. In plants, salicylate produced from isochorismate is important for the plant defense mechanisms known as local and systemic acquired resistance It has also been found to have secondary activity of catalyzing the transformation of chorismate to prephenate (chorismate mutase activity), although this is weak. The physiological role of IPL or PchB uses a pericyclic hydrogen transfer mechanism to produce salicylate from isochorismate as opposed to the notion that general base or acid catalysis would occur. This property resembles chorismate mutase enzyme that catalyze pericyclic reactions. IPL is a structural homolog of chorismate mutaselink title despite very low sequence identity (approx 20%). The similarity is restricted to the active sites which are conserved (Annu. Rev. Biophys. 2008. 37:153–73) and it has been found that chorismate mutase cannot be mutated to acquire IPL activity. IPL has been considered for improvement of its secondary activity computationally using hybrid quantum mechanics/ Molecular mechanics (J. AM. CHEM. SOC. 2008, 130, 2894-2895).Homologous genes are found in other microorganisms. It is also involved in bacterial siderophore synthesis. Peter Kast et al, propose for PchB a rare [1,5]-sigmatropic reaction mechanism that invokes electrostatic catalysis in analogy to the [3,3]-pericyclic rearrangement of chorismate in Chorismate mutases (CM).
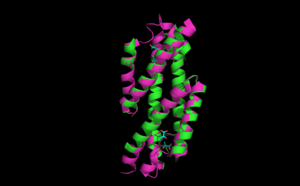
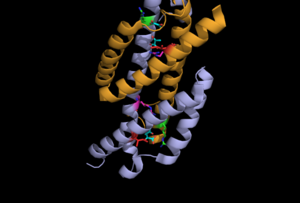
Superpositioning of the open and the closed PchB is shown to the right.
Structure
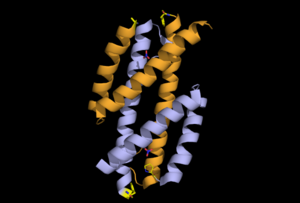
This is a 101-residue chain made of three alpha helices. It is found in the apo form, pyruvate bound open anf pyruvate bound closed forms.Some of the regions overlap with the regions of RdgC base domain which is a DNA binding recombination association protein.The quaternary structure of PchB was found to be dimeric as it is for EcCM (the homologous Chorismate Mutase), and most catalytic residues in the active site of EcCM are conserved in PchB. Moreover, it was shown that pchB complements the CM deficiency of an E. coli mutant strain and that PchB has low CM activity in vitro.
To the left is the apo form, center is the open pyruvate bound form and to the left is pyruvate bound closed form.
Chemistry
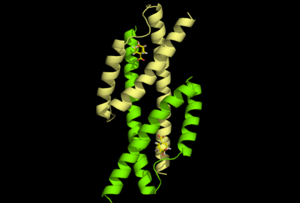
The transformation of isochorismate to prephenate is a pericyclic reaction. IPL catalyzes the elimination of the enolpyruvyl side chain from isochorismate to give salicylate and pyruvate. This type of aromatization reaction is generally formulated as a baseinitiated process, but a dissociative mechanism involving initial cleavage of the C-O bond to give an ion pair intermediate is conceivable. A concerted pericyclic pathway link title, in which the hydrogen atom at C2 is transferred to C9 of the side chain simultaneous with C-O cleavage, which represents a third possibility, has been proposed as well. The reaction is a one step mechanism (J. AM. CHEM. SOC. 2005, 127, 15002-15003).
To the right is the structure of 1ECM, homologue of pchB
Applications
Bacterial genes can be introduced into plants to increase the accumulation of the desired compounds. Salicylic acid content in Arabidopsis thaliana can be manipulated by expressing an engineered bacterial salicylate synthase by the fusion of two bacterial genes pchA and pchB from the human pathogen, Pseudomonas aeruginosa, that encode isochorismate synthase and isochorismate pyruvate lyase. These are expressed under a constitutive promoter.
Crystallization
X-ray crystallization is performed by hanging drop method for the apo, open and closed structures. The apo structure has nitrate in it in the regions that have the pyruvate in the bound structures. In the case of the bound structures, immediately prior to crystallization dilution in pyruvate is performed. These proteins carry the 6His Tag, but on crystallization, these are proteolyzed in the drop. Molecular replacement of the Apo with the structure of ECM (chorismate mutase) was done. Crystals of the bound structure are obtained by growing in PEG 3350. The resolution abtained for the Apo structure is 2.35 Angstroms and for the pyruvate bound structure it is 1.95 Angstroms. The residues in the loop that connects the first helix to the second has no residues lighting up in the data, depicting the disorderliness. These residues are present in case of the pyruvate bound structures in which case they are ordered.
| |||||||
Isochorismate-Pyruvate Lyase: apoenzyme to open conformation with pyruvate bound
made by chains A and B of 2h9c morphed to A and B of 2h9d
Click .
Click to see sidechains in the morph too.
Isochorismate-Pyruvate Lyase: open conformation with pyruvate bound morphing to closed conformation with pyruvate bound
made by chains A and B of 2h9d morphed to chains C and D
Click .
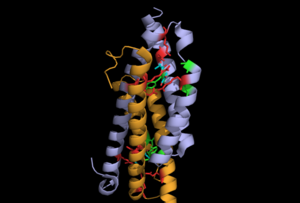
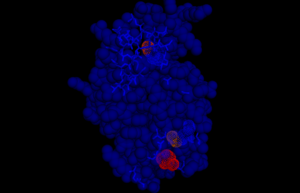
Active Site
The difference between the apo and the ligand bound open and close structures is in the ligand binding and the loop connecting the first and the second helix. In case of the open form, the loop is disordered and in case of the closed form it is ordered. The apo structure has nitrate ion in the active site (that is occupied by pyruvate) due to ammonium nitrate reservoir buffer during crystallization. In the pyruvate bound open and close structures, the residue that has an electrostatic interaction with pyruvate 1 is Arginine 31 and with pyruvate 2 it is Arg14. The nitrate in the apo structure forms an interaction with Arg31. It is observed that mutation of I87T would abolish the IPL activity.
Conserved Residues
5 amino acids that are conserved in the homologous protein 1ECM, which is a chorismate mutase are Arg14, Arg 31, Lys 42(disordered in the open structure), Arg 53 and Gln 90. There are 3 other residues that are not conserved in pchB, however (Tyr 86, Ala 50 and Val 54).
HotPatch analysis shows the active site as observed.
Correlation of structure with function
The pyruvates bound in the active site are correlated with the sidegroups involved in the reaction mechanism. Of the two pyruvates, one is generated by the reaction (Pyruvate 2 corresponds to the eolpyruvyl side chain cleaved in the reaction) interacting with ARg 14. Pyruvate 1, bound to Arg31 is in a position that would be occupied by the carboxylate group of the isochorismate substrate and the salicylate product. Wrong-ligand crystallographic refinement (WLCR) was done with the products of the reaction by Lamb et al and this satisfies with the catalytic mechanism.
Significance
This enzyme unlike others does not catalyze forming covalent enzyme-substrate intermediate or acid-base catalysis. Claisens rearrangement takes place, instead. The substrate is stabilized in an energitically unfavorable pseudoaxial conformation forcing the vanderwaals contact, producing steric strain on the substrate and provides electrostatic stabilization of the transition state. Lys 42 and Gln 90 are positioned to stabilize the developing negative charge of the polar transition product in the chorismate mutase reaction and the pyruvate byproduct in the IPL reaction. therefore the ordering of the active site loop shows induced fit of the enzyme upon substrate binding locks Lys 42 into place for catalysis.
3D structures of isochorismate pyruvate lyase
Updated on 07-July-2024
3log – MtIPL/isochorismate synthase - Mycobacterium tuberculosis
3rv6, 3rv7, 3rv8, 3rv9, 3st6, 3veh - MtIPL/isochorismate synthase + inhibitor
2h9c – PaIPL residues 1-99 – Pseudomonas aeruginosa
3hgw – PaIPL (mutant)
2h9d - PaIPL + pyruvate
3rem – PaIPL + pyruvate + salicylate
3hgx, 3ret – PaIPL (mutant) + pyruvate + salicylate
References
- ↑ DeClue MS, Baldridge KK, Kunzler DE, Kast P, Hilvert D. Isochorismate pyruvate lyase: a pericyclic reaction mechanism? J Am Chem Soc. 2005 Nov 2;127(43):15002-3. PMID:16248620 doi:http://dx.doi.org/10.1021/ja055871t
Additional Resources
For additional information, see: Amino Acid Synthesis & Metabolism
Proteopedia Page Contributors and Editors (what is this?)
Mangai Periasamy, Michal Harel, David Canner, Alexander Berchansky, Eran Hodis