Journal:Acta Cryst D:S205979832000501X
From Proteopedia

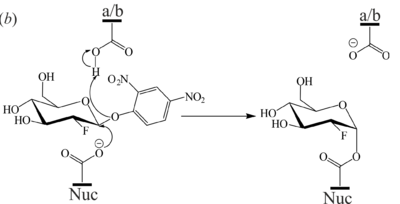
A Baculoviral System for the Production of Human β-Glucocerebrosidase Enables Atomic Resolution AnalysisRhianna J. Rowland, Liang Wu, Feng Liu and Gideon J. Davies [1] Molecular Tour Due to its involvement in GD, GBA is of considerable medical interest with continuous advances in the development of inhibitors, chaperones, and activity-based probes for therapeutic and diagnostic applications. The development of novel GBA inhibitors requires a source of active protein, however, the majority of structural and mechanistic studies on this enzyme rely on expired clinical formulations which are incredibly costly and difficult to obtain in sufficient quantities. This problem is further compounded by a lack of consensus on a reliable platform for GBA production for research purposes. There is thus a need for an alternative source of GBA to meet research demands and reduce the reliance upon clinical formulations. As part of our long-standing interest in the development of activity-based probes to study GBA and other glycosidases, we established a baculovirus-insect cell expression system for the production of GBA. The expression platform described herein produces active, crystallizable GBA which is comparable in enzymatic activity and biophysical properties to clinical preparations, rendering it. This protein crystallizes readily and is amenable to ligand-binding studies, as demonstrated by a novel structure in complex with the glucosidase inhibitor, 2,4-dinitrophenyl-2-deoxy-2-fluoro-β-D-glucopyranoside. Furthermore, a novel crystal form of GBA which diffracts to give a 0.98 Å unliganded structure was described. This is the highest resolution structure of GBA deposited to date, permitting exquisite atomic resolution analysis which reveals two conformations of the catalytic acid-base residue. studies. In light of its purity, stability, and activity, we envision that the baculoviral GBA production system described in this work will alleviate the over-reliance on clinical formulations, aid in future biochemical studies of GBA, and support structural-guided development of novel GBA ligands. (PDB 6tjk). N-glycans are shown in spacefill representation. . Domain I (residues 1-27 and 383-414) in red, domain II (residues 30-75 and 431-497) in blue, and domain III (residues 76-381 and 416-430) in gold. structure of bound bis-tris-propane which forms hydrogen bonds with Trp179, Asn234, Glu235, Glu340, Trp381 and an ethylene glycol (EDO) cryoprotectant molecules. (PDB 6tjj). . (PDB 6tjq). The 2F-Glc moiety is covalently bound to the catalytic nucleophile (Glu340) which occupies two conformations. a/b = catalytic acid-base, Nuc = catalytic nucleophile, EDO = ethylene glycol. (PDB 6tn1). Domain I (residues 1-27 and 383-414) in orange, domain II (residues 30-75 and 431-497) in violet and domain III (residues 76–381 and 416-430) in blue. N-glycans are shown in ball-and-stick representation. (PDB 6tjk). Red indicates areas of high RMSD between the protein backbones. Loop 1 contains residues 27-31, loop 2 comprises residues 314-319, and loop 3 contains residues 344-350. . A magnesium ion (peach) coordinated by four waters (grey), Glu340 (nuc) and Glu235 (a/b), occupies the active site. PDB references: Structure of Cerezyme at pH 4.6 6tjj; Crystal Structure of Recombinant GBA in Complex with Bis-Tris Propane 6tjk; Crystal Structure of Recombinant GBA in Complex with 2-Deoxy-2-fluoro-beta-D-glucopyranoside 6tjq; Unliganded Crystal Structure of Recombinant GBA 6tn1. References
| |||||||||||