A GH13 α-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides
Karan Wangpaiboon, Pasunee Laohawuttichai, Sun-Yong Kim, Tomoyuki Mori, Santhana
Nakapong, Rath Pichyangkura, Piamsook Pongsawasdi, Toshio Hakoshima and Kuakarun Krusong [1]
Molecular Tour
α-Glucosidase (E.C.3.2.1.20) is a carbohydrate-hydrolyzing enzyme, which generally cleaves α-1,4 glycosidic bonds of oligosaccharides and starch from the non-reducing ends. However, α-glucosidase from Weissella cibaria BBK-1 (WcAG) exhibited distinct hydrolysis activity against α-1,4 linkages of short chain oligosaccharides from the reducing end. It prefers to hydrolyse and , while it cannot hydrolyse cyclic oligosaccharides and polysaccharides. . Blue represents Domain A, whereas Domain B, C, and N are shown in yellow, red, and green, respectively. Calcium ion is in magenta.
The dimer formation of WcAG:
- .
- . Blue represents Domain A, whereas Domain B, C, and N are shown in yellow, red, and green, respectively. Lighter colors represent different subunit of dimer.
WcAG formed a , creating the substrate-binding groove. The residues near the dimer interface are shown in cyan and lavender spacefill representation, the maltotriose is shown in ball and stick representation and colored in yellow, the catalytic residues are colored magenta.
Ligand binding sites:
The carbon, nitrogen and oxygen atoms of the bound ligands are displayed in
cyan/yellow, blue and red, respectively. The three catalytic residues are shown in salmon sticks.
The active site of WcAG was naturally designed to fit perfectly with maltotriose. . E374/E374Q, D440, and D345/D345N are the catalytic residues (yellow) and other amino acid residues in/near the active site are shown in salmon. The asterisk and magenta colour present the residue from another subunit. The maltotriose is shown in cyan. White dashes represent hydrogen bonds.
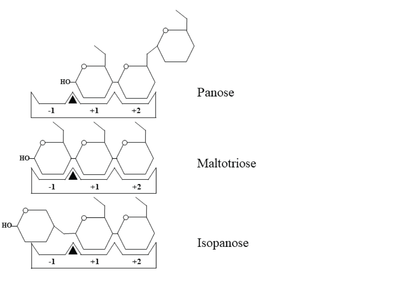
Schematic representation of panose, maltotriose and isopanose binding within the active site of
WcAG. The -1, +1, and +2 subsites are indicated as -1, +1, and +2, respectively. A black triangle represents the substrate cleavage site, located between subsites -1 and +1.
(grey) (PDB ID: 7ehi) is clearly observed by superimposition with maltotriose in WcAG structure (cyan) (PDB ID: 7dcg).
The in front of the active site modulates the substrate specificity of WcAG. This scene represents the movement of R176, E296 and F295 at two loops (P174-Y180 and T290-D300) in front of the active site when there is no substrate bound (grey) and when the active site is occupied by acarbose (salmon).
(PDB ID: 2d2o). The acabose is displayed in cyan, while yellow represents maltohexose. The residues E374Q, D345, and D440 are catalytic residues. The asterisk (*) indicates the residues from another subunit.
PDB references: α-glucosidase from Weisella cibaria BBK-1, wild type, 7d9b; E374Q mutant, complex with maltose, 7d9c; E374Q mutant, complex with maltotriose, 7dcg; E374Q mutant, complex with acarbose, 7dch; D345N mutant, complex with maltose, 7ehh; D345N mutant, covalent maltosyl-α-glucosidase intermediate, 7ehi.
References
- ↑ Wangpaiboon K, Laohawuttichai P, Kim SY, Mori T, Nakapong S, Pichyangkura R, Pongsawasdi P, Hakoshima T, Krusong K. A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides. Acta Crystallogr D Struct Biol. 2021 Aug 1;77(Pt 8):1064-1076. doi:, 10.1107/S205979832100677X. Epub 2021 Jul 29. PMID:34342279 doi:http://dx.doi.org/10.1107/S205979832100677X


