Journal:Acta Cryst D:S2059798322009755
From Proteopedia

Structural and functional investigation of the human snRNP assembly factor AAR2 in complex with the PRPF8 RNaseH domainMarco Preussner, Karine F. Santos, Jonathan Alles, Christina Heroven, Florian Heyd, Markus C. Wahl, Gert Weber [1] Molecular Tour (7pjh). In this and the following scenes: AAR2Δloop, orange; PRPF8RH, sky blue; a flexible loop (labeled Ser3)[2] of AAR2 connecting its two domains, which in AAR2Δloop was replaced by three serine residues and another smaller flexible loop between residues 313-321 are labeled. N- and C-termini as well as the β-finger module of PRPF8RH are labeled. in complex with human AAR2Δloop and yeast Aar2p/PRPF8JM (PDB ID 4i43)[3] respectively, to illustrate the human AAR2 in a larger PRPF8 context. In this and the following scenes: Aar2p, maroon; Prp8pRH, dark blue; Prp8pJM, cyan. . Water molecules are shown as red spheres. Interacting residues are shown as ball-and-sticks colored by atom type; carbon, as the respective protein; nitrogen, blue; oxygen, red; sulfur, yellow; dashed black lines, hydrogen bonds or salt bridges. . . . 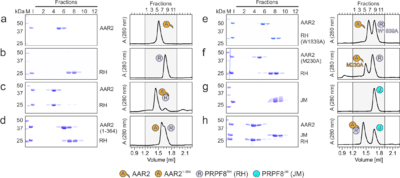 Figure 2. Probing AAR2Δloop-PRPF8RH interacting regions and residues. (a-h) SDS-PAGE analyses (left) and UV elution profiles (right) of analytical size exclusion chromatography runs monitoring the interactions among AAR2 variants, PRPF8RH variants and PRPF8JM. Figures a-c were adapted from (Santos et al., 2015)[2] and are shown for comparison. M, molecular mass standard (kDa); I, input samples. Protein bands are identified on the right. Elution fractions are indicated at the top of the gels and profiles, elution volumes are indicated at the bottom of the profiles. Icons are explained at the bottom. Variants are indicated at the respective icons. Peaks labeled by transparent icons represent an excess of the respective protein. 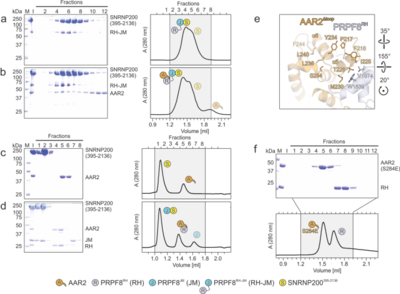 Figure 3. Probing AAR2Δloop-PRPF8-SNRNP200 interactions and AAR2 phosphorylation. (a-d) SDS-PAGE analyses (left) and UV elution profiles (right) of analytical size exclusion chromatography runs monitoring the interactions among AAR2, PRPF8RH-JM and SNRNP200395-2136 (a, b) and among AAR2, PRPF8RH, PRPF8JM and SNRNP200395-2136 (c, d). (e) Close-up view of the region in AAR2Δloop-PRPF8RH surrounding AAR2Δloop S284. The corresponding region in yeast Aar2p is profoundly restructured upon replacement of the equivalent S253 by a phospho-mimetic glutamate residue (Weber et al., 2013)[4]. (f) SDS-PAGE analysis (top) and UV elution profile (bottom) of an analytical size exclusion chromatography run monitoring the interaction between AAR2S284E and PRPF8RH. In panels showing SDS PAGE gels and elution profiles: M, molecular mass standard (kDa); I, input samples. Protein bands are identified on the right. Elution fractions are indicated at the top of the gels and profiles, elution volumes are indicated at the bottom of the profiles. Icons are explained at the bottom. Variants are indicated at the respective icons. Peaks labeled by transparent icons represent an excess of the respective protein. . The corresponding region in yeast Aar2p is profoundly restructured upon replacement of the equivalent S253 by a phospho-mimetic glutamate residue [4]. . Close-up views comparing the AAR2Δloop C-terminal tail (sticks) traversing the PRPF8RH domain below the protruding β-finger module (surface views):
The structure and functional data of the human spliceosomal assembly factor Aar2 in complex with a core spliceosomal domain of the PRPF8 protein indicates a different function of human Aar2 in contrast to the yeast protein. References
| |||||||||||
