Journal:Acta Cryst D:S205979832201186X
From Proteopedia

Structures of permuted halves of a modern ribose binding proteinFlorian Michel, Sooruban Shanmugaratnam, Sergio Romero-Romero, Birte Höcker [1] Molecular Tour The orientation of the two lobes to each other changes upon ligand binding, with a transition from a ligand-free open conformation to a ligand-bound closed one. This change in structure is often detected by associated receptor proteins and initiates specific downstream signaling processes. Among the diverse PBPs found in nature, the ribose binding protein (RBP) from Thermotoga maritima is a typical representative of the PBP-like type I fold and acts as a component for ribose transport. Despite diversity in the sequences of different PBP types, a shared common ancestry has been proposed. The structural symmetry of the two lobes has long led to the hypothesis that the PBP-like folds originated from a single-lobed ancestor protein, with a flavodoxin-like fold topology. To try and isolate a structural entity resembling the progenitor protein, we generated permuted halves of the N- and C-terminal lobes of RBP from T. maritima, purified, characterized, and solved their structures via X-ray crystallography. The obtained structures differ somewhat from the expected conformation, highlighting the inherent adaptability of this class of protein. This non-native structural rearrangement offers interesting clues for an evolutionary path for this protein fold. Tracing the mechanism of duplication of the single-lobed precursor, its adaptation to a bilobal form and their variations on a structural level can help us understand how this fold came about. 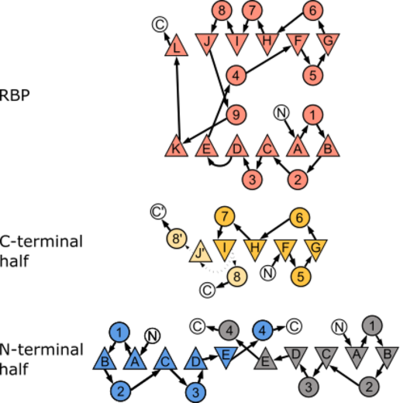 Canonical topology of both the parental RBP and the two halves investigated in this study. We observed an alternative arrangement of the halves compared to the one expected from the PBP-architecture. Figure adapted from Fukami-Kobayashi et al. (1999)[2] where β-sheets are depicted as triangles and α-helices as circles. Arrangement of the secondary structure elements reflect their three-dimensional order for RBP, RBP-CPC, and RBP-CPN. N- and C-termini are labeled with N and C, respectively, and the connections between the secondary structure elements are shown as arrows. The connection between β-strand “I” and its two possible configurations either to α-helix “8” or β-strand J’ in RBP-CPC are shown as dotted arrows as these stretches are not resolved in the crystal structure Cartoon representations of the structural alignments of (PFB id 7qsp; the edges of the unresolved region of chain A (D95 – M116) and B (A98 – M116) shown with spheres) and of (PFB id 7qsq). Differences in the topology are highlighted by comparison of the parental structure (salmon; PFB id 2fn9) with the solved crystal structures of (yellow) and (blue). Interestingly, the non-native conformation observed in RBP-CPN resembles the sequence of other types of PBPs found in nature. References
| |||||||||||
