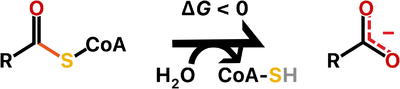Mitochondrial hotdog-fold thioesterase
From Proteopedia
Contents |
Overview of thioesterases
Thioesterases are enzymes that catalyze the hydrolysis of thioester bonds, which are the linkage between a carbonyl and a sulfur atom.
The ATP-dependent formation of a thioester bond from a carboxylate and a thiol in biomolecules makes them more reactive and is particularly an important commitment step in lipid metabolism. Therefore, thioesterases counteract this activation by releasing upon hydrolysis a molecule with the more stable carboxylate group. For this reason, thioesterases are found at the end of some metabolic pathways but they also may act as regulators of flux. Besides lipid metabolism, thioester bonds also occur in biosynthetic pathways for polyketide and non-ribosomal peptide production, as well as in main metabolites of carbon metabolism such as acetyl-CoA and succinyl-CoA.
There are two main families of thioesterases which are distinguished by their folding, named the α/β-hydrolases and the hotdog-fold hydrolases. Notably, these two different families are evolutionarily distant, so the thioesterase activity is a shared feature owing to convergent evolution.
| |||||||||||
Variable sequences within a conserved folding
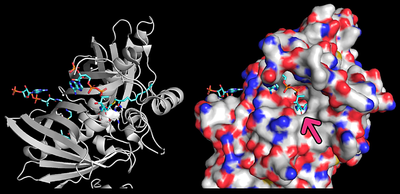
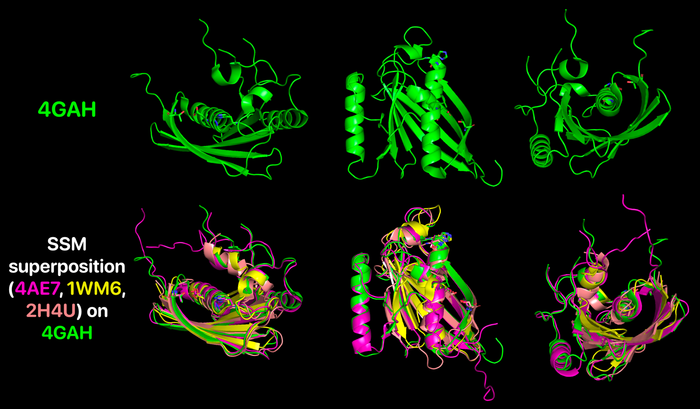
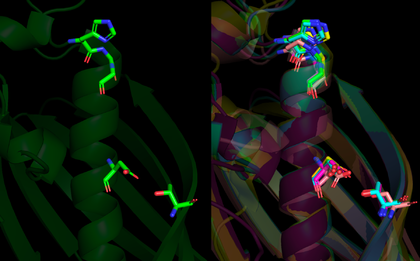
As mentioned previously, although the hotdog-fold thioesterases bear little similarity between their primary structure, they all share their characteristic conserved folding, which is exemplified by a comparison between the human paralogs Them4 (PDB 4GAH), Them5 (PDB 4AE7) and Them2 (PDB 2H4U) and the bacterial ortholog PaaI (PDB 1WM6) through a secondary-structure matching (SSM) superposition [3] using Coot [4] followed by spacial alignment using PyMOL. For this analysis, only the chain A from each structure is represented. It is clearly noticeable that their tertiary structures match precisely as the hotdog-fold domain.
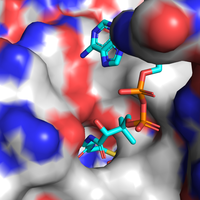
More specifically, by highlighting the residues from the HGG...D...T motif in Them4 and Them5 and its variants in Them2 and PaaI, it is observed that their orientations are also compatible, which is important for a putative binding of some acyl-CoA substrate in a similar way. Furthermore, the Them4 structure presented here was obtained in complex with the acyl-CoA structural analog undecan-2-one-CoA, whereas the other structures were obtained without a substrate nor such analog (the undecan-2-one-CoA molecules were omitted in this spacial alignment for a clearer comparison between the proteins's folding); although there is this difference, the residues from the catalytic motif in Them5, Them2 and PaaI crystallized without the ligand have a very similar orientation to the corresponding ones in Them4 with undecan-2-one-CoA. On one hand, this might suggest that the catalytic motif does not show a significant conformational change upon binding of the substrate; on the other hand, this might be a consequence of the fact that undecan-2-one-CoA is an structural analog of the acyl-CoA's substrates and not the acyl-CoA's themselves. In this latter case, in the presence of undecan-2-one-CoA, there might be a subtle difference upon binding of this molecule to the active site that does not cause a putative conformational change for catalysis.
Function
From enzymatic activities in vitro, it was shown that Them4[5] and Them5[6] have higher kcat/KM for acyl-CoA's with medium and long hydrocarbon chain, such as myristoyl-CoA (14:0), palmitoyl-CoA (16:0), oleoyl-CoA (18:1) and linoleoyl-CoA (18:2). According to Zhuravleva et al. (2012)[6], linoleoyl-CoA (18:2) was a preferred substrate for Them5. From studies with Them5−/− mice, it was identified by mass spectrometry (MS) that loss of Them5 is related to an increase in the levels of monolysocardiolipin (MLCL), which is a metabolite upstream of the cardiolipin remodeling process in mitochondria. Furthermore, the lipidomics analysis by MS for Them5−/− mice also revealed a 2-fold decrease of free fatty acids, notably linoleic (18:2) and linolenic (18:3) acids. This is consistent with the in vitro assay for the recombinant ∆34Them5 which revealed higher kcat/KM for linoleoyl-CoA (18:2). Moreover, it is observed by two-dimensional electron microscopy (2D-EM) and subsequent 3D reconstruction that in hepatocytes from Them5−/− mice, mitochondria were more elongated and interconnected, with a 2-fold increase in volume. With these data, Zhuravleva et al. (2012) [6] propose that Them5 might be a regulator of cardiolipin remodeling through modulation of the unsaturated acyl-CoA pool in mitochondria. This modulation in turn seems to affect mitochondrial morphology.
Zhao et al. (2012) [2] observed that Them4 shows very weak binding affinity (Ki > 1 mM) for carboxylic acids generated after the thioester bond hydrolysis, suggesting that this enzyme is not regulated by product inhibition.
Interestingly, it was revealed that Them2 interacts physically with the protein StarD2/PC-TP, which has a START (StAR-related lipid-transfer, with StAR standing for Steroidogenic Acute Regulatory protein) domain. More specifically, Kanno et al. (2007) [7] observed that StarD2 stimulates myristoyl-CoA hydrolysis catalyzed by Them2. It becomes even more curious when considering that Them1/ACOT11, another mammalian hotdog-fold thioesterase, has in its sequence an additional START domain.
Them4 is also called Akt Carboxyl-Terminal Modulator Protein (CTMP), owing to previous data suggesting that it interacts with the serine-threonine protein kinase Akt1 in an inferred mechanism of regulating apoptosis. At the plasma membrane, CTMP inhibits Akt by preventing its phosphorylation on key residues, threonine 308 and serine 473. This inhibition by CTMP reduces Akt's activity, which is crucial in insulin signaling, cellular survival, and transformation[8] . However, this activity is not well defined yet. Through pull-down assays, Zhao et al. (2012) [2] verified that Them4 and Akt1 form a stable complex and that Them4 inhibits Akt1 activity in vitro, but Akt1 does not inhibit Them4. In pathological contexts, such as skeletal muscle atrophy, CTMP has been shown to exacerbate muscle degeneration by reducing Akt signaling, thereby increasing muscle catabolism and decreasing protein synthesis[9] . In glioblastomas, CTMP expression is often downregulated due to promoter hypermethylation, removing its inhibitory effects on Akt and contributing to tumorigenesis[10] . Conversely, in head and neck squamous cell carcinoma (HNSCC), elevated CTMP levels enhance Akt phosphorylation, promoting tumor growth and metastasis, and correlate with poor prognosis[11] .
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Zhao H, Lim K, Choudry A, Latham JA, Pathak MC, Dominguez D, Luo L, Herzberg O, Dunaway-Mariano D. Correlation of Structure and Function in the Human Hotdog-fold Enzyme hTHEM4. Biochemistry. 2012 Aug 21;51(33):6490-2. Epub 2012 Aug 9. PMID:22871024 doi:10.1021/bi300968n
- ↑ Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004 Dec;60(Pt 12 Pt 1):2256-68. PMID:15572779 doi:10.1107/S0907444904026460
- ↑ Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004 Dec;60(Pt 12 Pt 1):2126-32. Epub 2004, Nov 26. PMID:15572765 doi:10.1107/S0907444904019158
- ↑ Zhao H, Martin BM, Bisoffi M, Dunaway-Mariano D. The Akt C-terminal modulator protein is an acyl-CoA thioesterase of the Hotdog-Fold family. Biochemistry. 2009 Jun 23;48(24):5507-9. doi: 10.1021/bi900710w. PMID:19453107 doi:http://dx.doi.org/10.1021/bi900710w
- ↑ 6.0 6.1 6.2 Zhuravleva E, Gut H, Hynx D, Marcellin D, Bleck CK, Genoud C, Cron P, Keusch JJ, Dummler B, Esposti MD, Hemmings BA. Acyl coenzyme a thioesterase them5/acot15 is involved in cardiolipin remodeling and Fatty liver development. Mol Cell Biol. 2012 Jul;32(14):2685-97. Epub 2012 May 14. PMID:22586271 doi:10.1128/MCB.00312-12
- ↑ Kanno K, Wu MK, Agate DS, Fanelli BJ, Wagle N, Scapa EF, Ukomadu C, Cohen DE. Interacting proteins dictate function of the minimal START domain phosphatidylcholine transfer protein/StarD2. J Biol Chem. 2007 Oct 19;282(42):30728-36. PMID:17704541 doi:10.1074/jbc.M703745200
- ↑ Maira SM, Galetic I, Brazil DP, Kaech S, Ingley E, Thelen M, Hemmings BA. Carboxyl-terminal modulator protein (CTMP), a negative regulator of PKB/Akt and v-Akt at the plasma membrane. Science. 2001 Oct 12;294(5541):374-80. PMID:11598301 doi:http://dx.doi.org/10.1126/science.1062030
- ↑ Wang J, Tierney L, Wilson C, Phillips V, Goldman L, Mumaw C, Muang E, Walker CL. Carboxyl-terminal modulator protein (CTMP) deficiency mitigates denervation-induced skeletal muscle atrophy. Biochem Biophys Res Commun. 2023 Feb 12;644:155-161. PMID:36652767 doi:10.1016/j.bbrc.2023.01.023
- ↑ Knobbe CB, Reifenberger J, Blaschke B, Reifenberger G. Hypermethylation and transcriptional downregulation of the carboxyl-terminal modulator protein gene in glioblastomas. J Natl Cancer Inst. 2004 Mar 17;96(6):483-6. PMID:15026474 doi:10.1093/jnci/djh064
- ↑ Chang JW, Jung SN, Kim JH, Shim GA, Park HS, Liu L, Kim JM, Park J, Koo BS. Carboxyl-Terminal Modulator Protein Positively Acts as an Oncogenic Driver in Head and Neck Squamous Cell Carcinoma via Regulating Akt phosphorylation. Sci Rep. 2016 Jun 22;6:28503. PMID:27328758 doi:10.1038/srep28503
Proteopedia Page Contributors and Editors (what is this?)
Marcelo Mesa, Thabata Fernanda Oliveira, Eduardo Ferraro, Michal Harel