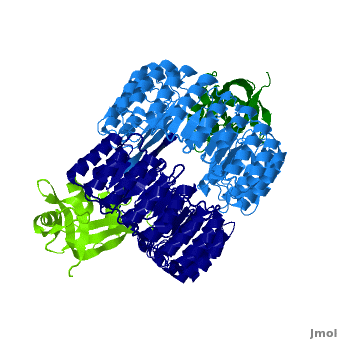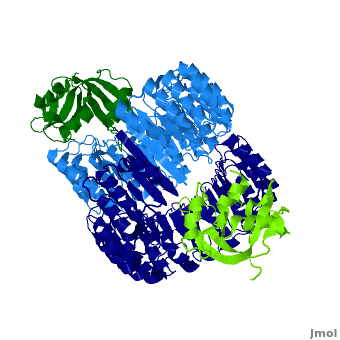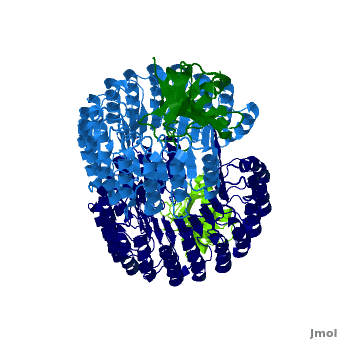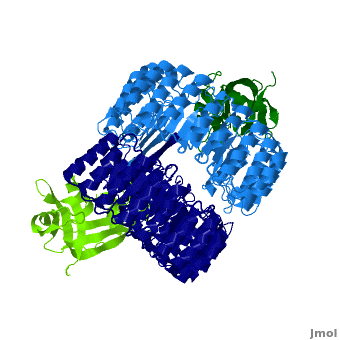
Function
Ribonuclease inhibitors (RI) or Ribonuclease/angiogenin inhibitor are a family of large (~450 residues, ~49 kDa), acidic (pI ~4.7), proteins that bind to and inhibit ribonucleases. Human RI(hRI) is a major cellular protein, comprising ~0.1% of all cellular protein by weight. [1]
Ribonucleases (RNase) are enzymes that degrade RNA and are often cytotoxic which gives them chemotherapeutic properties. However, when bound to an RI they are no longer functional. Understanding the mechanism through which RI identifies and binds to RNases will allow scientists to design/modify RNases to evade hRI. In fact, one drug, Onconase (ONC), a ribonuclease from the Northern Leopard Frog (Rana pipiens), is now in Phase III clinical trials as a cancer chemotherapeutic agent [2].
Structural Motifs
RI features 15 leucine-rich-repeats (LRR) alternating 28 and 29 residues. make up the outer circumference with on the inner circumference. There are no disulfide bonds in the While RI undergoes a conformation change upon binding of a substrate, there is no hinge. The lack of long-range stabilization allows for structural flexibility especially between the two ends of the molecule[3].
Interactions between hRI and RNase 1
The interaction between hRI and RNase 1 is one of the stronger known protein-protein interactions with a disassociation constant(Kd) of 10^-15 M [4].
hRI + RNase 1 ←→ hRI·RNase 1
Arg39 and Arg91 of RNase 1 are proposed to be “electrostatic targeting residues” a term used by Johnson et. al to define residues that push the formation of protein complexes[5] As shown, and form multiple hydrogen bonds to hRI, keeping the RNase in place, allowing the formation of salt bridges that further lock hRI and RNase together. The targeting residues hold the complex together while a total of nineteen intermolecular hydrogen bonds form. This includes a multitude of salt bridges which are especially strong.
Hydrogen Bonds:
RNase 1:::::::hRI
Arg4:::::::Trp438
Arg4:::::::Trp438
Lys7:::::::Ser460
Gln11:::::::Ser460
Arg31:::::::Gln10
Arg31:::::::Asp36
Arg32:::::::Asp36
Arg32:::::::Asp36
Arg39:::::::Glu401
Arg39:::::::Glu401
Arg39:::::::Glu401
Lys41:::::::Asp435
Asn67:::::::Tyr437
Asn71:::::::Tyr437
Asn88:::::::Glu264
Gly89:::::::Trp261
Arg91:::::::Glu287
Arg91:::::::Glu287
Glu111:::::::Tyr437.[6]
RI + RNase 1
Mechanism of Inhibition
One remarkable property of RI is the ability to recognize, bind, and inhibit different RNases that don't share common sequences or active sites. This next section will outline the mechanism by which RI inhibits the catalytic properties of RNase 1.
First, examine the of RNase 1. His12, Lys41, and His119 are three key catalytic residues to take note of. They have been colored CPK and represented in wireframe.[7] Tyr434, Asp435, Ser460 form hydrogen bonds to the citrate. the citrate forms hydrogen bonds with His12, Lys41, and His119 of RNase 1. The binding of citrate interferes with the substrate-binding pocket of RNase 1 severely affecting its enzymatic ability. [8]
Medical Implications
As mentioned earlier in the introduction, ribonucleases are cytotoxic. They bind to and chop up RNA. RNase is an endonuclease and is diffusion limited, meaning it acts as fast as susbstrates arrive. RNases exhibit great stability and are often purified by sulfuric acid treatment and then boiling until it is the only surviving macromolecule. RI’s exist to protect cells from rogue RNases. [9]
The amphibian RNase ONC is currently in clinical trials, however, human ribonucleases possess advantages over amphibian, namely increased catalytic ability, decreased renal toxicity, and decreased immunogenicity. The RI evasion of an RNase is critical to its chemotherapeutic effectiveness. Genetic engineers have been working on site-specific mutations that either decrease the association constant or increase the disassociation constant.
There are many interactions between hRI and RNase 1 providing a multitude of mutagenic options. However, it must be remembered that while changing residues can lead to increased RI evasion, because protein sequence determines structure, and structure determines form changing residues can also lead to decreased catalytic activity. Therefore, altering residues in exchange for RI evasion is a double-edged sword because cytotoxicity must be preserved. It's medically relevant to first understand the interactions between hRI and ribonucleases. Then one can design/modify a ribonuclease to be able to evade hRI while still maintaining functionality—potentially a new way to fight cancer. Researchers at the University of Wisconsin have been working on this very thing. One particular species, “R39D/N67D/N88A/ G89D/R91D RNase 1" has a 5×10^9-fold decrease in affinity for RI, while maintaing nearly wild-type ribonucleolytic activity, conformational stability, and cytotoxicity. [10] The R39D and R91D substitutions sever the hydrogen bonds formed by and that served as electrostatic targeting regions. The aspartates create negative/negative repulsion. This modified RNase provides an interesting example of the future direction and potential of biochemistry and medicine.
3D structures of RI
Updated on 08-August-2024
1a4y – hRI + RNase 5 – human
2bex – hRI + nonsecretory RNase
1z7x, 2q4g - hRI + RNase I
4peq - bRI + RNase I - bovine
2bnh - pRI - pig
1dfj – pRI + bRNase I
3tsr – RI + RNAse I – mouse
4per - RI + RNase I - chicken
References
- ↑ Shapiro R. Cytoplasmic ribonuclease inhibitor. Methods Enzymol. 2001;341:611-28. PMID:11582809
- ↑ Zwolinska M, Smolewski P. [Onconase: a ribonuclease with antitumor activity]. Postepy Hig Med Dosw (Online). 2010 Feb 19;64:58-66. PMID:20173221
- ↑ Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996 Dec 20;264(5):1028-43. PMID:9000628 doi:http://dx.doi.org/10.1006/jmbi.1996.0694
- ↑ Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007 Apr 27;368(2):434-49. Epub 2007 Feb 9. PMID:17350650 doi:10.1016/j.jmb.2007.02.005
- ↑ Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007 Apr 27;368(2):434-49. Epub 2007 Feb 9. PMID:17350650 doi:10.1016/j.jmb.2007.02.005
- ↑ Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007 Apr 27;368(2):434-49. Epub 2007 Feb 9. PMID:17350650 doi:10.1016/j.jmb.2007.02.005
- ↑ Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996 Dec 20;264(5):1028-43. PMID:9000628 doi:http://dx.doi.org/10.1006/jmbi.1996.0694
- ↑ Kobe B, Deisenhofer J. Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J Mol Biol. 1996 Dec 20;264(5):1028-43. PMID:9000628 doi:http://dx.doi.org/10.1006/jmbi.1996.0694
- ↑ http://www.pdb.org/pdb/101/motm.do?momID=105
- ↑ Johnson RJ, McCoy JG, Bingman CA, Phillips GN Jr, Raines RT. Inhibition of human pancreatic ribonuclease by the human ribonuclease inhibitor protein. J Mol Biol. 2007 Apr 27;368(2):434-49. Epub 2007 Feb 9. PMID:17350650 doi:10.1016/j.jmb.2007.02.005