Sandbox 206
From Proteopedia
|
In this article we want to present you a new YFP (Yellow Fluorescent Protein) with improved maturation and reduced environmental sensitivity due to 5 mutation of the well characterized variant of the YFP that is to say the EYFP (Enhanced Yellow Fluorescent Protein). The PDB number of this molecule is 1MYW. [1]
Contents |
A new YFP called Venus (1MYW)
Introduction
Yellow Fluorescent protein (YFP) is a variant of Green fluorescent protein (GFP). A lot of search was made of this protein in order to improve his fluorescence, his viability in different middle and others conditions[2]. This protein is very useful like a biomarker in a lot of biological applications. First, the GFP was mutated In YFP. But the YFP had a low fluorescence because of a slow maturation and it was very sensitive to experimental condition (pH, Cl-)[3]. To increase the brightness of the protein, maturation speed and other properties, five mutations were done to obtain Venus, a Yellow Fluorescent Protein less sensitive to experimental condition.
Presentation of the molecule
|
Primary and Secondary structure
Venus is a 27 kDa protein (26 875,50Da) consisting of 239 residues and form a secondary structure of 7 (2 in the center of the molecule) and 11 . This molecule is monomeric. Venus has many common point with the GFP and also with the EYFP. In all three structures compared, the cross-section taken perpendicularly to the long axis of the barrel is not perfectly circular but rather in shape. While the longer diameter of the oval cross-section of all three proteins is identical (23.8 Å measured from distances of four Cα pairs), the shorter diameter differs between EYFP (22.3 Å) and Venus (21.9 Å). The difference in the oval shape between these yellow variants is closely related to the packing of interior amino acid residues and influences the integrity of dimer interface[4].
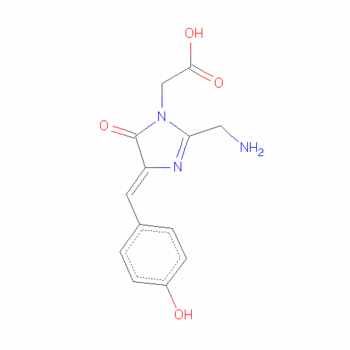
The Chromophore
Chromophores almost always arise in one of two forms: conjugated pi systems and metal complexes. In our case, the chromophore is a conjugated pi-bond system. In this type of chromophores, the electrons jump between energy levels that are extended pi orbitals, created by a series of alternating single and double bonds, often in aromatic systems.[5].
The residues surrounding the Venus are similar to those surrounding the chromophore of EYFP. Electron density studies of Venus show an 11° angle between the planes of the chromophore and Tyr203, while the same angle measured in EYFP is 11.5–12.3°, making the plane of the Venus chromophore slightly more parallel than that of EYFP.
This small difference may account for a minor change in the absorption spectrum of Venus rela- tive to EYFP, as seen at a pH range of 4.6–8.6 and a 50 mM NaCl concentration. Although the position of the main absorp- tion maximum remains unchanged (516 nm) in both Venus and EYFP, a smaller peak resulting from the neutral (protonated) chromophore is shifted in Venus by 20 nm producing a peak at 413 nm versus 393 nm in EYFP. The slightly more parallel orientation of the aromatic rings of Tyr66 and Tyr203 might also help improve the π-π interaction, thus reducing the excited state energy of the neutral chromophore. If this is the case, we can hypothesize that at lower pH, which is favorable to chro- mophore protonation, the angle between the rings would be smaller than 11°.
The different mutations
The five mutations are : F46L, F64L, M153T, V163A, and S175G F46L mutation rearranges some side chains near the chromophore. F64L induced conformational changes in the molecule. M153T, V163A, and S175G mutations improve the maturation by creating regions of greater flexibility.
Other Mutant of YFP
There is three YFP mutant that are used : our Citrine, venus and Ipet. You can also see on the GFP main page (here), many others Fluorescence Proteins (wild-types or mutants).
Applications
Venus can be used as a biomarker (YFP-tagged organelle marker for instance).
Thanks to Venus we can understand a lot of gene regulation mechanism. This protein is very useful in order to localise cancer cell for instance or different organelles. Indeed, venus can be used as a biomarker. Thank to its high brightness, the phenomena of transfer of energy can be directly displayed in the cell allowing to define at the same time the nature of proteins in interactions, the dynamics of these phenomena and their cellular localization.
We want to make you aware of one example to see concretely what could be the role of this molecule in some search. We decided to take the example of the Team IGEM 2008 of the Albert Ludwigs University of Freibourg (Germany). This is the site were you can found more informatrions
Abstract:
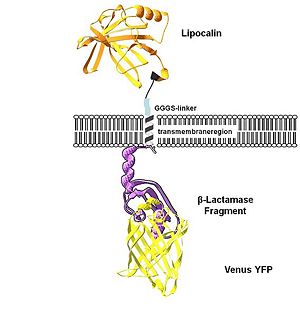
"Localization at the cell membrane
To show the localization of the constructs at the cell membrane transfection of the construct signal_peptide-Lipocalin-transmembrane_region-betaLactamase1-YFP was performed. Figure 2_Transfection shows the model of the protein structure based on PDB files. Anti-fluorescein Anticalin (BBa_K157004) is the extracellular part of the construct. The transmembrane region is identical to that of the EGF-receptor erbb1 (BBa_K157002). Split beta-Lactamase (BBa_I757011), the intracellular part is fused to the yellow fluorescent protein to detect membrane localization."
Experimental Details of the Crystallization
Method: X-RAY DIFFRACTION
Exp. Data: N/A
Resolution: 2.20 Å
R-Value: 0.218 (obs.)
R-Free: 0.248
Unit Cell :
| Length [Å] | Angles [Å] |
|---|---|
| a = 82.70 | α = 90.00 |
| b = 82.70 | β = 90.00 |
| c = 72.56 | γ = 120.00 |
This information is on the PDB page : here
Reference for this Structure
Rekas A, Alattia JR, Nagai T, Miyawaki A, Ikura M. Crystal structure of venus, a yellow fluorescent protein with improved maturation and reduced environmental sensitivity. J Biol Chem. 2002 Dec 27;277(52):50573-8. Epub 2002 Oct 4.
Pubmed : 12370172
DOI : 10.1074/jbc.M209524200
References
- ↑ http://www.rcsb.org/pdb/explore/explore.do?structureId=1MYW
- ↑ Improved green fluorescence; Heim R., Cubitt A.B. and Tsien R.Y; Nature 373, 663 - 664 (23 February 1995) Pubmed : 7854443 DOI : 10.1038/373663b0 Disponible on http://tsienlab.ucsd.edu/Publications/Heim%201995%20Nature%20-%20Improved%20GFP.PDF
- ↑ 1H, 15N and 13C assignments of yellow fluorescent protein (YFP) Venus; Hsu ST, Behrens C, Cabrita LD, Dobson CM.; Biomol NMR Assign. 2009 Jun;3(1):67-72. Epub 2009 Jan 13 Pubmed : 19636949 DOI : doi:10.1007/s12104-009-9143-y Disponible on https://springerlink3.metapress.com
- ↑ Crystal Structure of Venus, a Yellow Fluorescent Protein with Improved Maturation and Reduced Environmental Sensitivity; Rekas, A., Alattia, J.R., Nagai, T., Miyawaki, A., and Ikura M., 2002 Pubmed : 12370172 DOI : 10.1074/jbc.M209524200 Disponible on [1]
- ↑ http://www.chemguide.co.uk/analysis/uvvisible/theory.html#top
Additional Resources
Nikon : http://www.microscopyu.com/print/articles/livecellimaging/fpintro-print.html
Introduction to Fluorescent Proteins : http://www.microscopyu.com/articles/livecellimaging/fpintro.html
IGEM 2008 Uni Freiburg : http://2008.igem.org/Team:Freiburg
Proteopedia Page Contributors and Editors
Zoé Blanck, Camille Oms