Sandbox 719
From Proteopedia
|
In this article, we want to present you a red Fluorescent Protein (FP) recently cloned from a corallimorpharian of the Discosoma genus. The PDB number of this molecule is 1G7K [1]. The publications related is called "Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-Å resolution" [2]
Contents
|
The DsRed, a red fluorescent protein from coral
Introduction
Since the discovery of FP with the Green Fluorescents Protein (GFP) in the Aequorea victoria algae, other type of FP was discovered with differents colors such as cyan, yellow and red. All those proteins contribute to the natural coloration of their host and also permit the multicolor tagging experiments. The crystal structure of DsRed, has been determined at 2.0-Å resolution by multiple-wavelength anomalous dispersion and crystallographic refinement. In this article we want to present you the structure of the protein but also the function, use and a small comparison between de GFP and the DsRed.
Structure of the DsRed protein
Secondary Structure
|
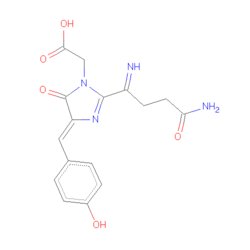
The DsRed is a 28kDa polypeptide that emits a red color after an excitation because of the presents of a chromophore composed by 3 amino acids –Gln-Tyr-Gly- (residues 66-68)[3]. The fold of our protein is very closed to the most common fluorescent protein, the Aequorea victoria Green Fluorescent Protein (avGFP). The primary sequence of those two proteins is 23% similar but in the immediate vicinity of the chromophore the primary structure is strictly conserved and theses several amino acid are probably essential for the chromophore formation. The DsRed protein is obligatory a tetramer in solution. The monomer is composed by 11-strand with a coaxial .
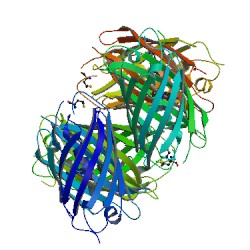
The tetramer Interface
The most striking feature of DsRed is that, as shown in Figure 1, it exists as an extremely close-packed tetramer, quite unlike the primarily monomeric avGFP. It is a dimer of dimer. The whole tetramer is a square prism with remarkably flat sides and a small elliptical hole directly through the center. The hole is lined with polar residues and salt bridges and localizes a number of solvent molecules. The accessible surface area of the isolated monomer is around 10,230 Ų, and that of the tetramer is 31,420 Ų, so roughly 25% of the monomer surface is not solvent accessible in the tetramer[4]. The AB interface does not seem to have particularly notable features and consists of hydrophobic interactions between small side chains, although a few hydrogen bonds and salt bridges are also present. On the other hand, interactions in the AC (and BD) interface consist largely of salt bridges and hydrogen bonds, many of which are mediated by buried water molecules, as well a surprisingly large number of interactions involving aromatic residues.
The Chromophore environment
|
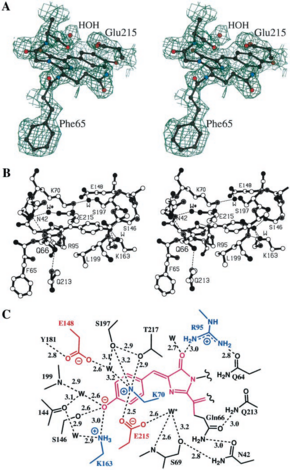
Structure of the chromophore was predicted by sequence comparisons. The chromophore of the DsRed is localized in a very polar cavity, and results to the autocatalytic cyclization and hydrogenation of the. Environment of DsRed chromophore is much more complicated in DsRd than in GFP, cause to the hydrogen bond and salt bridge network. There are more charges in DsRed than in GFP, due to three lysine (residues 70,83 and 163) and glutamic acid (residue 148) wich interact very closely with the chromophore. These charges are localized in two perpendicular band : the positive one is parallel to the axis of the chromophore, and the negative one perpendicular to this axis. The Phenolate oxygen of the chromophore forms a hydrogen bond with Ser-146 and a nearly water molecule, and it also forms a charge-charge with the Lys-163 residue. To forms all these interactions, the necessarily always charged. There is an other important residue in the struture of the chromophore, the residue Glu-215 (equivalent to the residue Glu-222 in GFP). This residue is highly close to the chromophore, and it also interacts with a water molecule localized near GLN-66. But Glu-215 also interacts with the positively charged chain of Lys-70, and it forms a hydrogen with Ser-205 side chain, like in GFP. The last notable interaction is the hydrogen bonding between Gln-213 and Asn-42, wich is probably important for the red-emitting fluorescence of the DsRed. The four chromophore in the tetramer form a rectangular array of 27-34 , suggesting that it exist a possible energy transfer between different chromophores. This could be important when we know the maturation of each chromophore have an efficient rate close to only 50 %.
Fluorescence
The broad excitation and emission bands have maxima at 558 and 583 nm, respectively (with a minor peak at 494 nm and a significant tryptophan peak at 280 nm) for a monomer extinction coefficient and fluorescence quantum yield at 558 nm of approximately 75,000 mol-1/cm-1 and 0.7, respectively[5].
Analisys and comparison between DsRed and GFP chromophore
These experiments shown the existence of a green fluorescent protein intermediate, very close to GFP, and suggests that there are several steps in the overall reaction (Matz et al.). Studies of Baird et al. shown there are two key conserved amino acids : Gln-66 and Gln-215. When we do the comparison of chromophore of DsRed and GFP, two points seems to be important. First Gln-66 of DsRed had a sp2 hybridization while Thr/Ser-65 (which are at the same place in the molecule) of GFP have a sp3 hybridization. Secondly, the conserved glutamate Glu-215 of DsRed is closer of the chromophore and is bonded to a water molecule close to GlN-66, which becomes oxidized. So this positionning of Gln-66 and Glu-215 should have a big influence in the red fluorescence, and Glu-215 could have two roles : In the formation of green fluorescence intermediate, and, by its environment, is crucial for formation of green or red emitting species. Matz et al. Suggested that GFP is a “broken” version of an ancestral red fluorescent protein, due to these obsevations.
Mutation studies
Several studies were lead on DsRed, by random and directed mutagenesis. These studies allowed to identify the influence of a certain number of amino acids on characteristics or on the 3D structur of the protein, and to understand the mechanism of the fluorophore. The replacement of Lys-70 by a Met (mutation K70M) lead to a green fluorescent protein, very close to GFP. The reason of this modification is that Lys residue interacts directly with the chromophore. In fact Lys-70 is certainly a charged residue and should interacts via a water molecule with the residues Glu-215 and Glu-148 of the chromophore. The replacement by the neutral residue Met make this interaction impossible. Mutation of Asn-42 (which should interacts with Gln-66) can be understood by the same way, and lead also to a green molecule. An other interesting mutation is the replacement of Lys-83. The effect of this mutation depend of the amino will replace the Lys. Actually, K83R and K83N mutations results in green fluorescent protein, but K83M substitution lead to latge redshift of absorbtion and emission maxima. In fact, substition with an Arg will increased the chain size, and so inhibite the maturation, but substitution with a Met (wich had mainly the same size than Lys, but different charge) change only absorbtion and emission behavior.
Advantage of the DsRed protein
In vivo, this protein obviously participate to the natural coloration of its host (in this Aequorea victoria), and it had a role in the protection against UV radiation. In vitro the DsRed can be used complementary to the famou Green flurescent protein GFP. Actually, Dsred absorbs in green at 558 nm and emitting a red fluorescente light at 583 nm. This wavelenght is very higher than all the other fluorescent protein, and allow to detect this red-emmiting easily in cytometry or fluorescent microscopy. Futhermore, in addtion to this use in multicolor tagging axperiments, this protein, and all red-emitting proteins, could be helpful to avoid self-fluorescence of the study design . They could also extand the range of resonance energy in transfert-based experiments (FRET). So in this application, pairs of fluorescent can be uses to detect proximity-related phenomena in vivo. On an other side, DsRed had also several drawbacks for its uses in laboratory :
- Its fluorescence level is very low, it means we need an high quantity of chromophores to detect it
- To be red-emitting, DsRed need a long time for it maturation, to forms the tetramer
- In a high quantity, it could cytotoxic and forms aggregates.
The aim of the studies on this protein is to understand how it work exactly, and find solutions to avoid these problems.
Experimental Details of the Crystallization
Method: X-RAY DIFFRACTION
Resolution: 2.0 Å
R-Value: 0.166 (obs.)
R-Free: 0.249
Unit Cell :
| Length [Å] | Angles [Å] |
|---|---|
| a = 56.37 | α = 90.00 |
| b = 112.27 | β = 93.81 |
| c = 70.69 | γ = 90.00 |
Informations are on this page
References
- ↑ http://www.rcsb.org/pdb/explore/explore.do?structureId=1G7K
- ↑ Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. PubMed : 11209050. PubMedCentral : PMC14609 DOI : 10.1073
- ↑ Fluorescent proteins from nonbioluminescent Anthozoa species. Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA.. PubMed : 10504696
- ↑ The interpretation of protein structures: estimation of static accessibility. Lee B, Richards FM. PubMed : here
- ↑ Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Baird GS, Zacharias DA, Tsien RY. PubMed : here
Proteopedia Page Contributors and Editors
Zoé Blanck, Quentin Demeyere