(RNase III, 2EZ6) is a class 1 RNase III hydrolase enzyme. In general its function, which seems to be universally conserved, is the to and cleavage of dsRNA. Endoribonuclease III appears to be crucial for an organisms ability to rapidly adapt to environmental changes through dsRNA processing and decay.
Universally, it has been shown to mediate RNA turnover at the post-transcriptional level through processing rRNAs, tRNAs, some mRNAs, as well as non-coding dsRNAs. In microbes, RNase III has been shown that it also represses the synthesis of virulence factors (through the cleavage of foreign RNA.) [1]
In eukaryotes, RNase III has also been shown to generate microRNAs and siRNAs, which are central to gene regulation pathways. Model organisms for RNase III include E. coli and Aquifex aeolicus, whose structures have been solved with x-ray crystallography. Below, the structures of RNase III from Aquifex aeolicus and Thermotoga maritima are used to study the structure and correlating function in more detail. These organisms RNase IIIs have been compared previously due to their structural similarity [2].
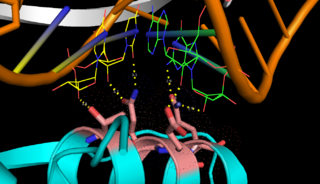
Fig.1.
RNase III binding to dsRNA Exploring the Structure
of Aquifex aeolicus RNase III (Aa-RNase III) are composed of an (endoND, green) and a (dsRBD, red)[3]. The crystal structure shows that RNase III is composed of a . The sequence of the endoND is characterized by a stretch of conserved residues (37ERLEFLGD44 in Aa-RNase III), which is known as the RNase III (purple) and makes up a large part of the active center, where catalysis takes place. RNase III hydrogen-bonds to dsRNA with , which make up the binding site (Fig.1). RNase III can affect gene expression in either of two ways: as a processing enzyme, where RNase III cleaves both natural and synthetic dsRNA into small duplex products averaging 9–18 base pairs in length, or as a binding protein which binds and stabilizes certain RNAs, thus suppressing the expression of certain genes[[4], [5]].
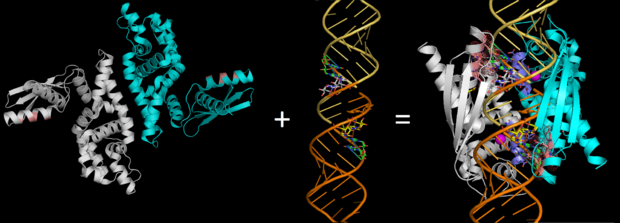
Fig.2.
RNase III (Pbd codes 1O0W & 2EZ6) conformation change when bound to dsRNA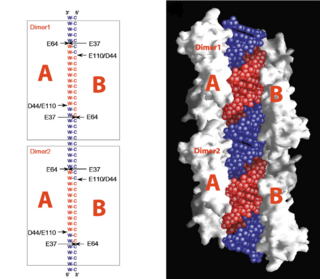
Fig.3.
cleavage site of Aa-RNase IIIOn the basis of the structural and biochemical data, catalytic models were proposed before the structure of a catalytic complex became available. Comparing the structure of Aa-RNase III with the structure of RNA-free Thermotoga maritima RNase III (RNA-free Tm-RNase III, PDB ID code 1O0W)[2] shows that there is and shift of dsRBD due to RNA binding, and there is a KGEMLFD between endoRD and dsRBD leading the rotation happened (Fig. 2). In addition, the two dsRBD domains are spaced apart from each other in a way that allows free rotation of dsRBD around the linker. In vivo data suggested that E110, E37, D44, and E64 are essential for catalysis[6]. This led to the model of the proteins active centers, which can accommodate a dsRNA substrate, each containing two different RNA cleavage sites, (pink and red). Specifically, E64 from each partner subunit, along with E37, E110, and D44 are located in the signature motif located at each end of the catalytic valley[7]. Fig. 3 presents a model showing Aa-RNase III cleaving dsRNA. This produces two identical RNA strands, each containing a 2 bp 3' overhang.
A Hypothetical Pathway Leading to the two Functional Forms of RNase III
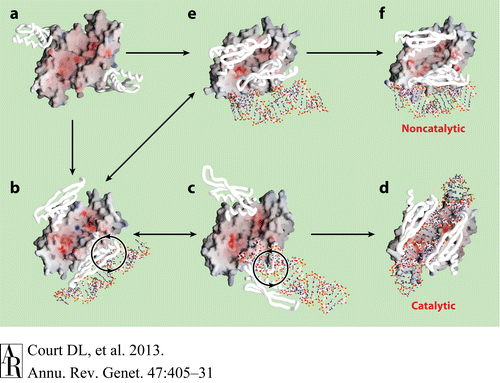
Fig.4.
Hypothetical Pathway Leading to the two Functional Forms of RNase IIIFig.4 presents a hypothetical pathway for RNase III to change from its inactive unbound state to two active dsRNA-bound states. The RNA-free RNase III dimer in conformation A (Figure a, PDB 1O0W) first binds a dsRNA with one of the two dsRBDs. The resulting complex may have at least two possible conformations, B (Figure b, PDB1YZ9 AaRNase IIIE110Q) and E (Figure e, PDB 1YYO), in which the dsRNA is located outside of the catalytic valley. These two conformations, b and e, appear to be interchangeable. In conformation E, the two dsRBDs pack against each other, hindering free rotation of the dsRBD-dsRNA complex around the flexible linker between the endoND and dsRBD. Conformation b allows free rotation of dsRBD-dsRNA around the linker (Figure b). The clockwise rotation of dsRBD-dsRNA leads to the catalytic form (Figure d, PDB 2EZ6) via conformation C (Figure c, PDB 1YYW). Conformations B and C are also likely to be interchangeable. In contrast, a factor that uncouples binding and processing can further stabilize conformation E leading to the noncatalytic form of RNase III (Figure f, PDB 1RC7)[[6], [8], [2]].




