This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1123
From Proteopedia
Contents |
Introduction
Since 2000, 38,1 million people were infected by the human immunodeficiency virus (HIV)[1]. This virus can cause the acquired immunodeficiency symptom (AIDS) which can often be lethal. The HIV belongs to the retrovirus family, more precisely to the lentivirus. This virus infects immune cells (T cells, macrophages, …) by integrating its genome in the host cell genome. Since HIV is a RNA virus, it converts its RNA into DNA thanks to the reverse transcriptase. Nevertheless, when its genome is integrated in the host genome, it can stay in latency during a long time.
The virus becomes active when the cell starts to fight another infection. The host cell transcript its own DNA and the viral DNA. This viral DNA codes for lytic proteins and viral proteins. The lytic proteins will kill the host cell, thus releasing the new viral particules. Indeed, it is the immune cells death which induces the decrease of the immune response and enhances thus the sensibility to other infections. Moreover, the viral proteins will produce new viruses which will be released and infect new cells. In this way, the knowledge of the different viral proteins, like the capsid proteins, can allows us to find new targets in order to avoid the viral dispersion/propagation ? [2]
Function
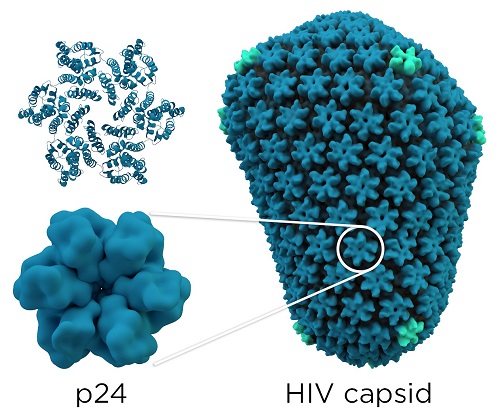
The HIV capsid is also called CA (for capsid) or p24. Its main role is to contain and protect the viral genome (RNA) and some enzymes (reverse transcriptase, integrase and protease). These proteins are indispensable because they are responsible of the structural assembly and organization of the virion. Indeed, p24 can interact with other viral proteins to assure the virion assembly and the RNA releasing. Furthermore, it can interact with host cell proteins during the viral cycle. [3]
Structural highlights
|
|
Structural basis of HIV-1 mature capsid
Each HIV-1 virus owns a conical core capsid that encapsulates the ssRNA(+) viral genome and some viral enzymes that are essential for the host infection. This capsid contains about 250 hexamers and 12 pentamers of the CA protein, pentamers are important to generate the curvature of the structure. This capsid is the mature capsid, its formation results in the maturation and disassembly of the immature gag polyproteins (first structural protein products which are encoded by the HIV-1 genome) by viral proteases. CA proteins are organized in two domains: an N-terminal domain ( ) and a C-terminal domain (). The mature capsid is formed by the assembly of approximately two-thirds of the mature CA proteins in the viral particle. CA proteins bind together and organize themselves in order to create a lattice of hexameric rings, these rings contain an inner ring of six s, surrounded by a belt of six s. Then, 12 CA pentamers join the structure, it permits the formation of a closed protein shell. [4]
Primary structure
CA proteins are composed of about 230 residues [5] and as we mentioned above they contain two domains: (in green) and (in yellow). These two domains are linked together thanks to a (in orange). [4]
Tertiary structure
CA tertiary protein structure is composed of seven helices in the NTD domain and eight helices in the CTD domain. The conserved tertiary structures take different quaternary arrangement during the new viral particle assembly. [6]
Quaternary structure
CA hexamers formation is due to intermolecular highly cooperative and noncovalent interactions, between the NTD of one subunit and the CTD of the neighbouring subunit within the same hexameric ring. These NTD-CTD interactions are crucial for the HIV-1 capsid assembly. CTD-CTD contacts and NTD-NTD contacts also exist for CA hexamers and pentamers formation. [4]
NTD forms both hexameric or pentameric rings, while CTD forms symmetric homodimers which connect the rings into a hexagonal lattice. Moreover, the formation of hexamers and pentamers is controlled by an electrostatic switch, thanks to this process, hexamers are favored compared to pentamers. Pentamer and hexamer structures are very close to each other, in either case, the CTDs form a belt which encircles the inner ring of NTDs.
Intermolecular NTD-NTD contacts facilitate the formation of the NTD rings, while NTD-CTD contacts maintain the CTD subunits in the belts. There is no intramolecular interaction between the NTD and CTD of each subunit, except the peptidic linkage between these two domains of course. Furthermore, the flexible linker is able to adopt different conformations, which is very useful because the monomers can position themselves in an optimal manner to permit the interaction surfaces in pentamer and hexamer. NTD rings form a rigid structure, this is not the case of CTDs in belts which are mobile and able to rotate, relative to the NTD ring. CTD pivotes such as a rigid body about four intermolecular helix-capping hydrogen bounds at the NTD-CTD interface.
Thus, each hexameric ring can have slightly different dihedral angles relative to its adjacent rings thanks to the CTDs movments. NTD rings interactions are possible thanks to the first three α-helices of each subunit, they forme a 15-helix barrel in the pentamer and a 18-helix barrel in the hexamer. A small hydrophobic core is located at the center of the bundle thanks to the presence of aliphatic residues, while polar sidechains are present at the periphery and do hydrophilic interactions. [6] [7]
Interactions with others partners
Even if p24 is classified as a structural protein, it is also involved in many cellular infection processes.
You can find bellow in a non exhaustive list of p24 partners :
- Cyclophylin A [10]
- Nuclear pore (certainly but not sure)[11]
- Dynein [11]
- CPSF6, TNPO3, NUP538/RanBP2 and nucleoporin 153 kDa (all these proteins are cellular transport factors)[4]
- Integrase ?
The HIV-1 capsid acts like a kind of "nuclear localisation signal" because it targets directly the virus toward the nucleus, where the integration takes place.
Capsid as therapeutical target
|
Thank to its central role in viral infectious process (genome protection, enveloppe cohesion, uncoating, budding), HIV capsid is a very good target for antiviral drugs. As a result, many research teams are working in this way and some molecules such as [11] are very promising. By binding to a "central pocket" on P24, it was shown in vitro that this compound is inhibiting the new viruses assembly, and consequently the virus budding.
This approach is interesting because, based on structural information, we are able to build such ligands using drug design. However, we are still far away from the "miraculous HIV drug", because the pathway from design to approved drug is not an easy way at all.
References
- ↑ Global HIV and AIDS statistics
- ↑ Weiss RA (May 1993). "How does HIV cause AIDS?". Science 260 (5112): 1273–9.Bibcode:1993 Science 260.1273W. doi:10.1126/science.8493571. PMID 8493571.
- ↑ [http://onlinelibrary.wiley.com/doi/10.1111/j.1742-4658.2009.07315.x/full The capsid protein of human immunodeficiency virus: interactions of HIV-1 capsid with host protein factors Authors : Anjali P.Mascarenhas, Karin Musier-Forsyth First published: 8 October 2009DOI: 10.1111/j.1742-4658.2009.07315.x]
- ↑ 4.0 4.1 4.2 4.3 [http://www.ncbi.nlm.nih.gov/pubmed/?term=%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BC%EF%BF%BCStructural+basis+of+HIV-1+capsid+recognition+by+PF74+and+CPSF6 Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):18625-30. doi: 10.1073/pnas.1419945112. Epub 2014 Dec 17. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Bhattacharya A, Alam SL, Fricke T, Zadrozny K, Sedzicki J, Taylor AB, Demeler B, Pornillos O, Ganser-Pornillos BK, Diaz-Griffero F, Ivanov DN, Yeager M.]
- ↑ PDB File: 3H47
- ↑ 6.0 6.1 [http://www.ncbi.nlm.nih.gov/pubmed/?term=Structure+of+the+imma+%CC%8Ature+HIV-1+capsid+in+intact+virus+particles+at+8.8+A+resolution Nature. 2015 Jan 22;517(7535):505-8. doi: 10.1038/nature13838. Epub 2014 Nov 2. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Schur FK, Hagen WJ, Rumlová M, Ruml T, Müller B, Kräusslich HG, Briggs JA]
- ↑ Nature. 2011 Jan 20;469(7330):424-7. doi: 10.1038/nature09640. Atomic-level modelling of the HIV capsid. Pornillos O, Ganser-Pornillos BK, Yeager M.
- ↑ Fernandez J, Gärtner K, Becker A, et al. HIV-1 capsid interacts with cytoskeletal-associated proteins for intracytoplasmic routing to the nucleus. Retrovirology. 2013;10(Suppl 1):P34. doi:10.1186/1742-4690-10-S1-P34.
- ↑ Fernandez J, Portilho DM, Danckaert A, Munier S, Becker A, Roux P, Zambo A, Shorte S, Jacob Y, Vidalain PO, Charneau P, Clavel F, Arhel NJ. Microtubule-associated proteins 1 (MAP1) promote human immunodeficiency virus type I (HIV-1) intracytoplasmic routing to the nucleus. J Biol Chem. 2015 Feb 20;290(8):4631-46. doi: 10.1074/jbc.M114.613133. Epub 2014 Dec 11.
- ↑ [http://jvi.asm.org/content/84/10/5181.long Marisa S. Briones, Charles W. Dobard and Samson A. Chow. Role of Human Immunodeficiency Virus Type 1 Integrase in Uncoating of the Viral Core. Accepted manuscript posted online 10 March 2010, doi:.1128/JVI.02382-09 J. Virol. May 2010 vol. 84 no. 10 5181-5190]
- ↑ 11.0 11.1 11.2 HIV-1 capsid: the multifaceted key player in HIV-1 infection Nature Reviews Microbiology 13,471–483 (2015)doi:10.1038/nrmicro3503
Structural capsid image : By Thomas Splettstoesser (www.scistyle.com) (Own work) [CC BY-SA 4.0 (http://creativecommons.org/licenses/by-sa/4.0)], via Wikimedia Commons