Sandbox Reserved 1654
From Proteopedia
| This Sandbox is Reserved from 26/11/2020, through 26/11/2021 for use in the course "Structural Biology" taught by Bruno Kieffer at the University of Strasbourg, ESBS. This reservation includes Sandbox Reserved 1643 through Sandbox Reserved 1664. |
To get started:
More help: Help:Editing |
Contents |
Cytoplasmic Polyadenylation Element-Binding Protein (CPEB)
CPEB (Cytoplasmic polyadenylation element binding protein) is present in most vertebrates and invertebrates and can activate or inhibit translation[1]. In human body there are 4 different isoforms of CPEB (CPEB1, which has a length of 65 amino acids, to CPEB4), distributed throughout body in a tissue-dependent manner and which interact differently with mRNA[2]. CPEB protein regulates the length of the polyA tail which allows to control the translation. It binds to mRNA and in association with some factors, can act as a translational repressor or activator, depending on these factors.
Structure
All CPEB proteins have a similar structure :
- A N-terminal region which is a regulatory region with phosphorylation and dephosphorylation sites. This region is variable in length and composition.
- A C-terminal region, composed of 2 recognition patterns : RRMs domains and zinc finger domains.
| Total Structure Weight | |||||||||||
| Atom Count | 543 | ||||||||||
| Number of protein chains | 1 | ||||||||||
| |||||||||||
About 54 residues with 6 cysteines and 2 histidines involved in a bond with a zinc atom, conserved for all isoforms and species. The modification of one of the eight zinc ligands destabilize the connection to the mRNA. includes :
- A (Rd turn, residues 515-520), which is stabilized by hydrogen bonds between amide and sulfure.
- β-hairpin with (residues 525-527) and (residues 533-535) between which there is an helical turn stabilized by hydrogen bonds.
- An (residues 538-545) which forms the second bridge between the two zinc-binding sites. The surface-exposed face of the helix has a potential for specific intermolecular interactions with nucleic acids or proteins. Therefore, it is this area that would be a platform to bind different proteins (ePAB, PARN, ...) by making hydrogen bonds.
- A 310 (residues 550-552).
- 2 zinc binding sites, the first one is composed of and the second is composed of .
- RRMs patterns[4] [2]
| Total Structure Weight | |||||||||||
| Atom Count | 1786 | ||||||||||
| Number of protein chains | 1 | Number of nucleic acid chains | 1 | </tr>||||||||
| |||||||||||
RRMs are necessary and sufficient for the CPE sequence recognition on RNA. They bind to RNA with high affinity and allow the RNA to take the good position. RRM1 binds to the four first RNA nucleotides (UUUU) and RRM2 binds to the 3' adenine of CPE. The two RRMs take a V-shaped conformation, facing to each other:
- has anti-parallel beta strands between the and the .
- The takes a helical turn that interacts with residues of the N-terminal extension and with .
- positions RRM2 relative to RRM1 by inserting between the beta sheet and of RRM2.
- After the helical turn, the interdomain linker folds in a which is anti-parallel to the (RRM2). The interdomain linker is therefore a kind of joint for the relative orientation of the two RRMs.
The N-terminal region of CPEB includes residues in the fourth β strand of the RRM2 domain. Within the linker region between RRM2 and ZZ domains, Leu510 shows long-range interactions with the aromatic ring of Tyr535 in the β2 strand, suggesting that there may be a close interaction between these two domains of CPEB meaning a close interaction between RRM2 and ZZ domains. Disruption of the CPEB-ZZ domain structure could affect the stability of the RRM2 domain structure through loss of the interdomain interface.
Function
General function
CPEB controls the balance between senescence and proliferation. Indeed, due to its two structural domains in its C-terminal region, it has the capacity to modify mRNA[2]. RRMs domains allow binding to the CPE sequence of mRNA (pyrimidine rich : UUUUUAU), thus ensuring good RNA positioning and high fidelity. Zing finger domains allow binding to different proteins, which play a role in affinity but not in specificity. It's therefore its ability to recruit different proteins that will determine its action, activation or repression of translation.
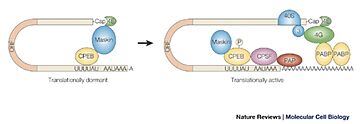
A specific arrangement of CPEs in mRNA can lead to the repression of the translation. In this case, the CPEB can form a dimer which could avoid the bound of the polyA polymerase complex in 2 different ways. It could prevent the association of ePAB with the polyA tail because CPEB recruits the deadenylase PARN which reduce the length of the polyA tail. It could disrupt the interaction between the binding factors of the translation eIF4E and eIF4G too, because CPEB recruits the protein Maskin which blocks eIF4G recruitment too[2]. This prevents the bound of the cap machinery to the mRNA and therefore inhibits the translation.
On the other hand, the CPEB can activate the translation. Indeed, in the cytoplasm, there are some repressed or silenced mRNA with a short polyA tail. They can be activated by cytoplasmic polyadenylation thanks to a hormonal stimulation. This stimulation can lead to the phosphorylation of CPEB which increase its affinity with the CPSF (Cleavage and Polyadenylation Specificity Factor) and decrease the binding between CPEB and PARN. CPSF binds to the mRNA at the sequence 3’ of the tail of the mRNA (AAUAAA) and recruits the poly(A) polymerase which leads to the elongation of the polyA tail and therefore to the activation of the translation[2].
Function in memory
Long-term and short-term memories differ by the duration of their retention. Long-term memory formation needs transcription and translation of stored mRNAs. CPEB operate in the post synaptic domain of neurons. CPEB is stimulated by a neuronal stimulation, it “activates translation of CaMKII and similar mRNAs. It also associates with motor proteins for mRNA transport and has a role in packaging of bound mRNAs to RNP complexes. CPEB initiates polyadenylation induced translation of dormant mRNAs during Xenopus oocyte maturation. In developing oocytes, following nuclear export, CPE containing mRNAs are bound by CPEB1 as well as other interacting proteins like PARN (poly A ribonuclease) and Gld2 (polyA polymerase), leading to removal of polyA tail of mRNAs, as PARN overrides Gld2 activity. This leads to translational suppression of mRNAs. However upon activity-induced phosphorylation of CPEB, PARN dissociates from the complex and the mRNA is polyadenylated leading to translation.” [3]
Diseases
CPEB proteins play a key role in some diseases, especially in cancers. Indeed, in some humans tumors the level of CPEB 1 is lower than in healthy cells and this leads to the growth of these tumors. An overexpression of CPEB 4 can lead to tumor growth too. Some researches try to find a cancer treatment thanks to CPEB mutations[2].
Other diseases, like Fragile X syndrome, could be treated by regulating the expression of CPEB. Indeed, this disease is due to a mutation on FRM1 gene, which is bind to X chromosome. Because of this mutation, FMRP, which is a translational repressor protein, isn’t expressed. So, proteins are overexpressed and it could be the cause of some dysfunctions observed for this disease. That’s why the regulation of the level of CPEB could treat this syndrome[5].
References
- ↑ Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007 Jun;32(6):279-85. doi: 10.1016/j.tibs.2007.04.004. Epub , 2007 May 4. PMID:17481902 doi:http://dx.doi.org/10.1016/j.tibs.2007.04.004
- ↑ 2.0 2.1 2.2 2.3 2.4 Fernandez-Miranda G, Mendez R. The CPEB-family of proteins, translational control in senescence and cancer. Ageing Res Rev. 2012 Sep;11(4):460-72. doi: 10.1016/j.arr.2012.03.004. Epub 2012 , Apr 21. PMID:22542725 doi:http://dx.doi.org/10.1016/j.arr.2012.03.004
- ↑ Merkel DJ, Wells SB, Hilburn BC, Elazzouzi F, Perez-Alvarado GC, Lee BM. The C-Terminal Region of Cytoplasmic Polyadenylation Element Binding Protein Is a ZZ Domain with Potential for Protein-Protein Interactions. J Mol Biol. 2013 Mar 13. pii: S0022-2836(13)00156-3. doi:, 10.1016/j.jmb.2013.03.009. PMID:23500490 doi:10.1016/j.jmb.2013.03.009
- ↑ Afroz T, Skrisovska L, Belloc E, Guillen-Boixet J, Mendez R, Allain FH. A fly trap mechanism provides sequence-specific RNA recognition by CPEB proteins. Genes Dev. 2014 Jul 1;28(13):1498-514. doi: 10.1101/gad.241133.114. PMID:24990967 doi:http://dx.doi.org/10.1101/gad.241133.114
- ↑ Udagawa T, Farny NG, Jakovcevski M, Kaphzan H, Alarcon JM, Anilkumar S, Ivshina M, Hurt JA, Nagaoka K, Nalavadi VC, Lorenz LJ, Bassell GJ, Akbarian S, Chattarji S, Klann E, Richter JD. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013 Nov;19(11):1473-7. doi: 10.1038/nm.3353. Epub 2013 Oct 20. PMID:24141422 doi:http://dx.doi.org/10.1038/nm.3353