This is a default text for your page Sandbox 1677. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function of your protein
Inositol polyphosphate 1-Phosphatase found in the organism Bos Taurus, is an enzyme that removes a phosphate group from a substrate through hydrolase. Specifically, the phosphomonoester bond is removed from position 1 phosphate of Ins(1,4)P2 or Ins(1,3,4)P3 to yield inositol 4-phosphate or inositol 3,4-bisphosphate. Another function this enzyme does is the regulation of gluconeogenesis and nucleotide metabolism within organisms.
Biological relevance and broader implications
This enzyme is important for this organism, Bos Taurus because it not only does inositol signaling but regulation of gluconeogenesis and nucleotide metabolism which are a couple of important pathways that help the organism function. Not only is INPP1 in cattle but can be found in humans and mice with possible slight variances in the function of the enzyme, though the article just picked one of the many organisms that have this enzyme. However, studying this organism is relevant because it allows us to study lithium-inhibited enzyme structures and functions to better understand them Also, possibly further our knowledge on cellular communication networks and/or cellular pathways (metabolisms) on cellular and molecular levels. This could result in advancements in human medicine and understanding enzymes in the human body on a molecular level on how they work.
Important amino acids
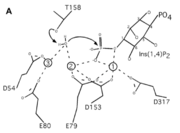
INPP1D54A does not have a catalytic triad though it has six important (D54, E80, E79, D153, D317, T158) that promote catalysis in the reaction along with the metal ions that helped the protein function. These amino acids hold the metal ions in place and metal ions hold on to the water and substrate. Thr158 is a very important catalytic amino acid because it is going to do a nucleophilic attack on the water molecule (shown in the picture on the right) which will result in a cascade of other reactions to happen (series of bonds breaking and being made) eventually leading to a phosphate leaving group. Hence, why the function of the enzyme is to remove a phosphate group.
Inositol (1,4)-bisphosphate is a that binds in the active site of INPP1D54A. The phosphate (1-PO4) in this substrate serves as a ligand for both the calcium ions in the mutated version of INPP1. [3] The substrate is held in place by certain (Glu269, Gly290, Lys294, Ala291, Ser157, Thr158) that play an extensive role in interactions such as hydrogen and ionic bonds between 1-PO4 and 2-PO4 of the substrate to the active site of the enzyme. Also, there are a few different ones that interact with the inositol ring and help reinforce the binding of the Ins(1,4)P2 by possible hydrophobic, hydrogen, and ionic bonds.
Structural highlights
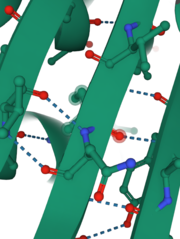
INPP1D54A contains 13 of which 10 are alpha helices and 3 are beta-sheets. Percentage-wise, this enzyme is 77% alpha-helices, 23% beta-sheets. Alpha helix 8 and beta-sheet 2 each contain one catalytic amino acid. Also, helix 6 and 7 and beta-sheet 2 form important interactions with the substrate (have 6 of the 9 amino acids that interact with the substrate). These alpha-helices and two large anti-parallel beta-sheets help determine the structure and function of this protein. Also, they provide stabilization through hydrogen bonding (shown in the picture on the right) that solidifies the conformation of beta-sheets and helices. This protein is already folded in its which is colored coated to show polar (pink) and hydrophobic (grey) amino acids indicating potential intermolecular interactions such as hydrophobic interaction, disulfide bridge, ionic, and hydrogen bonds. In addition, INPP1D54A and INPP1 do not exhibit a quaternary structure since it only consists of one subunit rather than two or more subunits.
A molecular model of INPP1D54A displays how much space the atoms within the protein occupy and gives the relative dimensions and shape of the protein. It also shows some of the (red) that interact with the protein and play an important part in the nucleophilic attack on the phosphate group removed from as the substrate binds to the active site. This image with the substrate transparent gives insight into the pocket where the substrate binds to the active site on the enzyme.
Other important features
In the structure INPP1D54A, there is a mutation in the amino acid aspartic acid (D)54 and causes it to change to alanine (A)54 (). This mutation does not impact the substrate affinity but does decrease the activity of INPP1. [3]
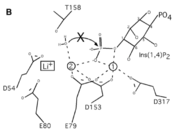
A six amino acid , DPIDxT anchors the metal-binding sites in the protein that are likely involved in catalysis while the metal binds to the substrate. [3] The sixth amino acid, x, is not as important as the other five, however, it can be any amino acid depending on the related crystallized structure to INPP1D54A or to a similar protein. Also, lithium is an uncompetitive inhibitor for this enzyme and when it binds to a metal site (metal site 3 in the lower image B) it causes the enzyme not to function properly.