Introduction
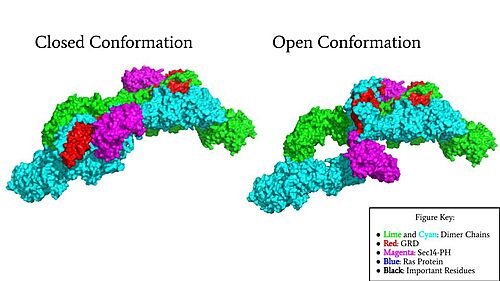
Figure 1: Surface Rendering of Neurofibromin in its Open (7PGT) and Closed (7PGR) Conformation.
Neurofibromin functions as a tumor suppressor protein by associating with Ras, a protein that signals for cell proliferation and growth. It has two conformations, the open, active conformation and the closed, inactive conformation (Figure 1). [1] The change in conformation of Neurofibromin from open to closed regulates Ras activity, turning it off, thus stopping cell growth from continuing in excess. Neurofibromin is a dimer made up of two identical monomers consisting of the Sec 14-PH domain and the GRD (Gap-related domain). The orientation of these domains is important for the interaction and binding of Ras to the Arginine finger in Neurofibromin, a key interaction in the regulation of Ras activity. Neurofibromin in its open conformation allows for the Ras association, unlike the closed conformation which is unable to associate with Ras and turn it off. [2] The closed conformation is stabilized by a triad consisting of Cys1032, His1558, and His1576 and a Zinc atom. The molecular structure and detail of Neurofibromin have been determined by Cryo-Electron Microscopy. Additionally, the structure of Neurofibromin isoform 2 revealed different functional states for Neurofibromin. [2] Neurofibromin is encoded by the NF1 gene, which is located on chromosome 17. Mutations in the NF1 gene are associated with diseases including Neurofibromatosis Type 1, Plexiform Neurofibromas, and cancers including glioblastoma, neuroblastoma, lung, ovarian, and breast cancer. [3] [4] [5]
Function
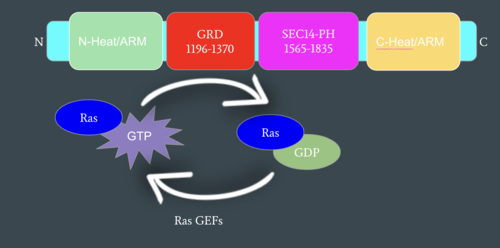
Figure 2: Mechanism of Ras Regulation by Neurofibromin. One chain of the homodimer is represented in cyan with its important domains highlighted. Ras Guanine Nucleotide Exchange Factors (RasGEFs) are shown catalyzing the transition from an active to an inactive Ras molecule and Neurofibromin is shown catalyzing the reverse reaction
Neurofibromin functions as a tumor suppressor protein.[1] It prevents cell growth by turning off Ras which in its active state, stimulates cell growth and division (Figure 2). Ras is a small, monomeric GTPase that binds to GTP and hydrolyzes it to GDP. When Ras is active, it subsequently activates other proteins that stimulate cell growth and proliferation. Ras is membrane-bound and interacts with Neurofibromin, a cytoplasmic protein, in the open conformation of Neurofibromin. Neurofibromin is brought to the membrane to associate with Ras by SPRED1. SPRED1 is a protein that helps regulate the Ras/MAPK signaling pathway responsible for the growth and proliferation of cells. Binding of SPRED1 to Raf in the MAPK signaling pathway blocks the activation of Raf, halts the rest of the pathway, and stops cell growth and proliferation. Unlike Ras, Neurofibromin can interact with SPRED1 in both the open and closed conformations.[2] The interaction between Neurofibromin and Ras is activated via an Arginine finger (Arg1276) present in the GRD domain of Neurofibromin. Arg1276 is only accessible for binding when the GRD and Sec14-PH domains are rotated into the open conformation. Ras binds to the arginine finger of Neurofibromin with its switch regions 1 and 2.[1] When Arg1276 is able to associate with Ras, Neurofibromin downregulates the Ras signaling pathway by speeding up Ras's GTPase activity, hydrolyzing the GTP associated with Ras to GDP. In its GDP bound state, Ras is inactive and cell growth and division is inhibited. [2] The positively charged arginine finger stabilizes the transition state for GTP hydrolysis by neutralizing the negative charges on GTP, which helps increase the speed of hydrolysis, fulfilling its catalytic function. [1]
Structure

Figure 3: Neurofibromin Important Domains in one chain of the homodimer
Neurofibromin is a made up of two identical chains. Neurofibromin has two conformations, open and closed. Shifting between these controls neurofibromin's ability to associate with Ras and perform its function of Ras regulation. The transformation between the overall closed and open conformations transitions it from an active to inactive state. There are involved in the transition between the open and closed conformations, the domain and the domain. The GRD and the Sec14-PH domain are centrally linked by an asymmetric, homodimeric core of four ARM repeats and 27 HEAT repeats (Figure 3). The GRD and Sec14-PH domains extend out from the core and then return to the core. The orientation of the GRD and Sec-14 in relation to the N-HEAT/ARM and C-Heat/ARM determine what conformation neurofibromin is in. Although neurofibromin is a homodimer with two identical protomers, only one protomer has its GRD and Sec14-PH domains rotated into the .[2]
Conformational States
Closed Conformation
In the , the GRD and Sec14-PH domains are rotated so Ras cannot bind. In this conformation, the GRD and Sec14-PH are inaccessible and inactive. Neurofibromin is held in the inactive state by a consisting of residues Cys1032, His1558, and His1576 that form a transition metal-binding site with zinc. The rigid organization of the keeps the GRD domain packed tightly on top of the Heat Arms in the Neurofibromin core. This tight compaction sterically occludes Neurofibromin from In its active form, Ras and Neurofibromin will associate via an (Arg1276). However, the from the Neurofibromin core in the closed conformation inhibits this association. Therefore, in the closed conformation, neurofibromin cannot catalyze GTP hydrolysis by Ras and Ras continues to signal for cell growth and proliferation.[2]
Open Conformation
In the , the GRD and Sec14-PH domain on one protomer have rotated and become accessible for binding to Ras. This transition is initiated by the movement of the transition metal-binding site. The Cys1032, His1558, and His1576 Residues become separated and zinc is not able to bond. In the active form, one protomer has its GRD and Sec14-PH domains oriented oppositely from the inactive form (Figure 4). The GRD rotates -130° and the Sec14-PH domain rotates -90° away from the N-HEAT/ARM. Due to this rotation, Cys1032 is now located too far away, approximately 30 Å, from His1558 and His1576 which results in the loss of the metal-binding site and no formation of the . The lack of the transition metal-binding site allows the GRD to orient itself to [2]. This association positions the to help stabilize and orient a catalytic Ras residue (Q61) so that the gamma phosphate of GTP can be nucleophilically attacked [6]. When Neurofibromin is in the open, active conformation, is able to bind to Ras because there is no steric hindrance from the Neurofibromin core.[2]
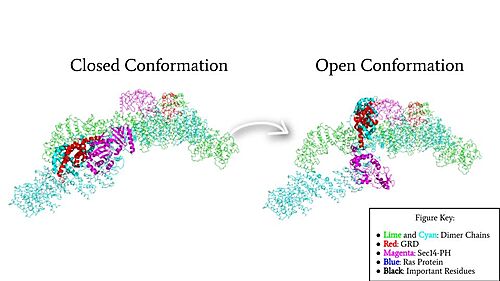
Figure 4: Rotation of the GRD and Sec14-PH domains from the closed conformation (7PGR) of neurofibromin to the open conformation (7PGT) of neurofibromin to allow Ras binding. The GRD rotates -130° and the Sec14-PH domain rotates -90°
Disease and Medical Relevance
Currently over 1485 mutations of have been identified that lead to life-threatening illnesses or conditions such as Neurofibromatosis Type I.[3] [5] Mutations to the NF1 gene that prevents the interaction of Neurofibromin with Ras, remove this check on Ras-dependent cell proliferation and uncontrolled cell growth can lead to tumors and a higher risk of cancer. Neurofibromatosis Type 1, the most well-known disease resulting from mutations in Neurofibromin, is characterized by cognitive impairment, soft, non-cancerous tumors on or under the skin known as neurofibromas, birthmarks called cafe-au-lait macules, clusters of freckles in unusual places, and problems with the bones, eyes and nervous system.[4]
Neurofibromin is also involved in the differentiation of neural crest-derived cells, mesenchymal cells, neural cells, melanocytes, and bone cells. As Neurofibromin is essential for embryonic development, mutations to the NF1 gene can result in psychological retardation resulting from Type I neurofibromatosis. Most of the 1485 point mutations identified to lead to a synthesis of truncated, non-functional protein. Type I Neurofibromatosis is inherited in an autosomal dominant manner but about 50% of cases result from de novo mutations. [5]
Mutations in the NF1 gene have been found in sporadic cancers such as glioblastoma, neuroblastoma, lung cancer, ovarian cancer, and breast cancer. Researchers are still unsure as to whether biallelic loss of NF1 is common or if it is only a hemizygous loss of the gene that contributes to the growth progression of certain sporadic tumors.[4]
Student Contributors
- Hannah Luchinski
- Sophie Mullinix




