Sandbox Reserved 1847
From Proteopedia
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Contents |
Structure
Introduction
What are the SARS-CoV2 Minibinders?
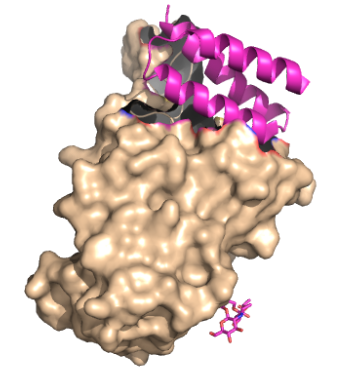
These mini proteins target the interaction between ACE2 and SARS-CoV-2 spike protein [1]. The mini binders are small proteins designed to bind to the SARS-CoV-2 spike protein with a greater affinity than ACE2 [1]. These mini binders reduce the viral burden of SARS-CoV-2 in mice [2]. Minibinders were de novo designed to mimic the ACE2 helix, but have a lower dissociation constant, yielding a greater affinity for the spike protein [1]. The binding region between (PDB: 8K4U) makes it seem like binding region is telling how these proteins were designed. Taking a closer look at the SARS-CoV-2 disease pathway shows where the minibinders target the interaction between ACE2 and SARS-CoV-2 spike protein.
SARS-CoV-2 Disease Pathway
Minibinders mimic the pathway of the SARS-CoV-2 virus attachment to the cell surface receptors. In the standard process, the SARS-CoV-2 surface spike proteins ACE2. This anchors to the host for viral entry [3]. When the spike protein binds to the ACE2 receptor, the cell membrane-associated protease, TMPRSS2, activates the spike protein, promoting viral entry [4]. The activated spike protein then cleaves itself into S1 and S2 subunits [4]. The S1 subunit contains a receptor binding domain that recognizes and binds to ACE2 [4]. The S2 subunit undergoes a conformational change which permits viral entry [4]. The S2 domain has a fusion peptide (FP) domain that will help regulate membrane fusion by disrupting and connecting the host cell’s membrane [4]. The S2 domain is also composed of HR1 and HR2 subunits, which are heptapeptide sequences involved in the entry of SARS-CoV-2. HR1 is located at the C-terminal domain of a hydrophobic FP, and HR2 is located at the N-terminal of the transmembrane domain [4]. In this conformational change, the S2 subunit inserts its FP domain into the host cell's membrane. Once the host cell’s membrane is penetrated, this triggers an interaction with the HR2 domain and HR1 trimer, forming the 6-helical bundle. The bundle brings the viral envelope and cell membrane in close enough distance for viral fusion and ultimately viral entry [4]. Once inside, the virus translates viral proteins, eliciting an immune response and spreading the viral particles throughout the body [4].
See also: Coronavirus Disease 2019 (COVID-19)
Minibinders block SARS- CoV-2 Acceptor Binding
The primary goal of the mini binders is to prevent the spike proteins from binding to ACE2, blocking viral membrane attachment [2]. Because the mini binders have a greater binding affinity than ACE2 for the spike protein, they prevent viral entry and infection [5]. Targeting this specific interaction between the SARS-CoV-2 spike protein and ACE2 are potential targets for future therapeutics to treat the virus [4]. There is a demand for more treatments for SARS-CoV-2. These minibinders pose an advantage to other therapeutics, such as antibodies, because they are much smaller in size and more stable [1]. As for vaccines, constantly updating and modifying them drains finances and time. The faster and more cost-appropriate answer is in the minibinders [1].
Expectations of this page
The focus of this page is on the design of mini binders and how they effectively prevent the entry of the viral SARS-CoV-2 spike protein. To explain inhibition by the minibinders, we will initially describe the structure of the spike protein and its interaction with ACE2. We will then explain the helical design of minibinders and how they block spike protein function. Laying the foundation for the mini binders, we will then take a look at how the mini binders are designed to obtain the best possible helical structure. With that, then it is finally time to look at the receptor binding domain between the various mini binders and their interactions with the spike protein.
Design
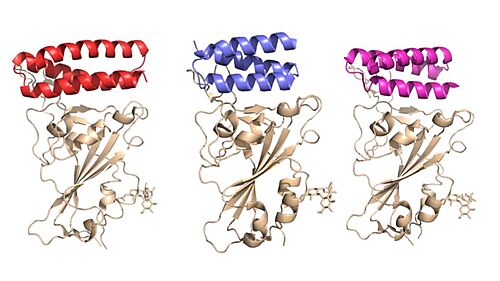
AHB2 is a Series A Helix Binder which is a miniprotein that binds specifically to the alpha helix of the protein. LCB1 and LCB3 are long chain bases that are longer in length which allow for more contact between the RBD, increasing the binding affinity. These mini binders, AHB2 and LCB1, were designed de novo with the intention to mimic the binding of ACE2 to the spike protein [1]. Using Rotamer Interaction Field (RIF) docking, the proteins were able to make the most efficient bonding using the ACE2 spike protein binding interface [1]. Using Site Saturation Mutagenesis (SSM), every residue in the minibinder’s helix scaffold was substituted with each of the 20 amino acids, one at a time, computationally choosing the best sequence [6]. Forming SSM libraries,experimental tests were run on each of the libraries to converge on a small number of closely related sequences [6]. From these libraries, one of these was selected for each design, AHB2 or LCB1-LCB8[1]. AHB2 was designed using an ACE2 helix scaffold, while LCB1 and LCB3 were designed completely from scratch, attempting to make the best possible helix with the greatest affinity for the spike protein receptors . Although LCB1 was designed before LCB3, LCB3 was less effective at neutralizing the viral response with a high IC50 Value [1] . These minibinders are small proteins, modeled similarly to the ACE2 and SARS-CoV-2 spike protein. There were two strategies utilized. One strategy included directly incorporating the ACE2 helix of the RBD and creating more interactions, increasing the binding affinity of the minibinders [1]. The other strategy was designing the minibinders completely from scratch, completely dependent on the RBD [1]. AHB2 utilized the first method, incorporating the ACE2 helix, while LCB1 and LCB3 utilized the second method [1] .
Implications
Potency of the minibinders
Examining the IC50 values of the various mini binders gives quantitative data to the effectiveness of the proteins in preventing an immune response. The highest IC50 was AHB2 (15.5 nM), followed by LCB3 (40.1 pM) LCB1 (23.5 pM) [1]. The higher IC50 indicates a larger concentration of mini binder required to inhibit the biological process. Both LCB1 and LCB3 proved to be significantly more effective than AHB2, LCB1 and LCB3 were within 3-fold of the most potent anti-Spike monoclonal antibodies described to date [1].
Results from mice study
The effectiveness of the most potent minibinder was examined in mice. LCB1 was administered to the mice via nasal delivery. As expected, compared to control mini protein, the LCB1 was significantly more effective at reducing the viral burden, diminishing the immune cell infiltration, and inflammation [2]. The virus was not detected in the lungs 4-7 days post-infection, and the spleen, heart, and brain had viral RNA at very low concentrations [2].
Benefits of minibinders over other therapeutics
The size of these mini binders is a large reason why they are so effective. The minibinders have a 20-fold more potential for nebulization compared with antibodies, and the molecular weight of the minibinders is 5% of a full antibody molecule [1]. When the LCB1 wasa attached to a human IgG domain to enhance bioavailability, staying in the body longer, LCB1 was less effective [2]. This is likely due to the increase in size when bound to the antibody. The high stability of the mini binders allows them to be administered as a gel via nebulization [1]. Future directions of mini binders are to increase the efficiency of the process to obtain a sequence for pathogen neutralizing designs more promptly [1]. Given that there are only a small number of antibody therapies and vaccines approved for treatment of SARS-CoV-2, minibinders as potential therapeutics may lay the foundation for similar minibinders designs as treatments for other viruses.
| |||||||||||
|