Sandbox Reserved 1851
From Proteopedia
| This Sandbox is Reserved from March 18 through September 1, 2025 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson and Mark Macbeth at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1828 through Sandbox Reserved 1846. |
To get started:
More help: Help:Editing |
Contents |
Novel Diels-Alder Catalyst Identified Using de novo Design
|
Introduction
General Structure
Active Site
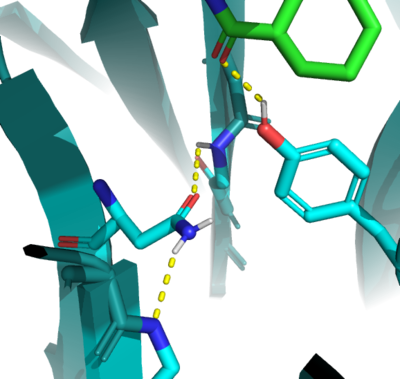
In the active state, there are two catalytic that aim to stabilize the transition state of the Diels-Alder reaction. Y134 acts as a to the oxygen on the . Q208 acts as a to the nitrogen on the ligand as well as a donor to the neighboring oxygen. These interactions help reduce the energetic gap between orbitals allowing the reaction to proceed, outlined in HOMO/LUMO.
Helix Addition
In the evolution process, researchers added a 16-residue to the top of the binding site. The hydrophobic helix “functions as a lid to constrain the substrates in a productive orientation for reaction” (CITE), decreasing the Km of the enzyme and increasing the catalytic efficiency, as seen in the measured kinetics of the enzyme.
Mechanism
Uncatalyzed Reaction
Orbital Stabilization
Structural Details
Active Site
Development & Evolution
DA_20_10
DA_20_10 provided key mutations in and around the active site that increased the hydrophobicity, provided structural stability, and increased interactions between the ligand and surrounding residues.
Q162R
Residue 162 resides near the top of the binding entrance to the enzyme, and is within 3A in most models on the enzyme. It can act as a hydrogen bond donor to the terminal phosphate on the ligand when in proximity. To increase this interaction, the group chose a Q to R mutation, which decreased the length of the potential hydrogen bond to within 2.5A, increasing the strength of the interaction.
S284A
Residue 284 resides deep within the binding pocket of the enzyme. The group chose an to mutation to increase the hydrophobicity of the binding pocket and reduce reactivity, without also changing any steric characteristics in the region unintentionally near the catalytic residues.
A285N
Residue 285, as follows, is also buried within the binding pocket. The group introduced this mutation to increase steric hindrance with the catalytic tyrosine, reducing the number of rotamers the residue has to increase the reactivity of the enzyme by lowering the distance between Y134 and the ligand.
Relevance
Chemical Applications
Improvements
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.
</StructureSection>
References
Student Collaborators
Micah Zile Kate Thuma Taylor Donahue