Sandbox Reserved 468
From Proteopedia
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||||||||||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
Matrix Metalloproteinase-1 (MMP-1)
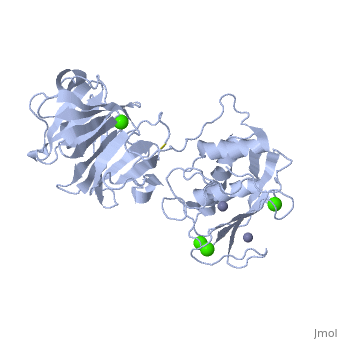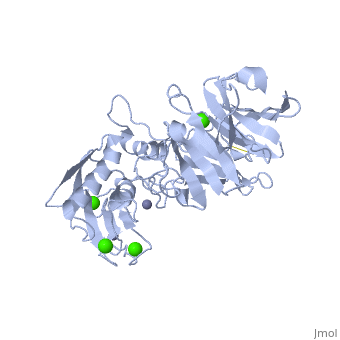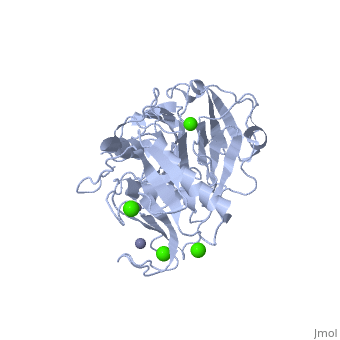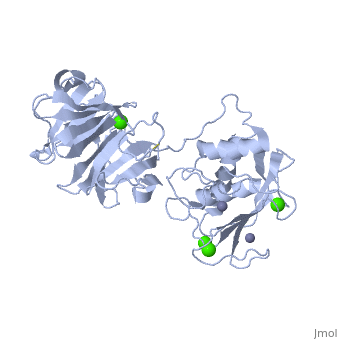
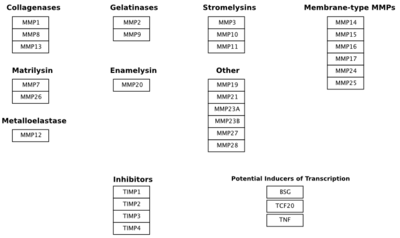
Matrix Metalloproteinase-1 (MMP-1)is a collagenase made up of two monomers. Collagenases are enzymes that break down the bonds in collagen [1]. MMP-1 in humans is encoded by the MMP1 gene. Interestingly, MMP-1 was actually the first vertebrate collagenase that was both purified as a protein and cloned as cDNA [1]. MMP-1 belongs to a family of enzymes known as Matrix metalloproteinases (MMPs). These enzymes are known as zinc-dependent because of the zinc ions found in their catalytic sites. The MMPs belong to a larger family of proteins known as the metzincin superfamily. MMPs can degrade all kinds of extracellular matrix proteins as well as a number of other bioactive molecules [2]. They are known to be involved in the cleavage of cell surface receptors, the release of apoptotic ligands (such as the FAS ligand), and chemokine/cytokine in/activation. MMPs are also thought to play an important role in cell behaviors such as cell proliferation, migration (adhesion/dispersion), differentiation, angiogenesis, apoptosis,host defense, embryonic development, reproduction, and tissue remodeling. MMPs are also involved in disease processes, such as arthritis and metastasis [2]. This image shows the entire MMP family.
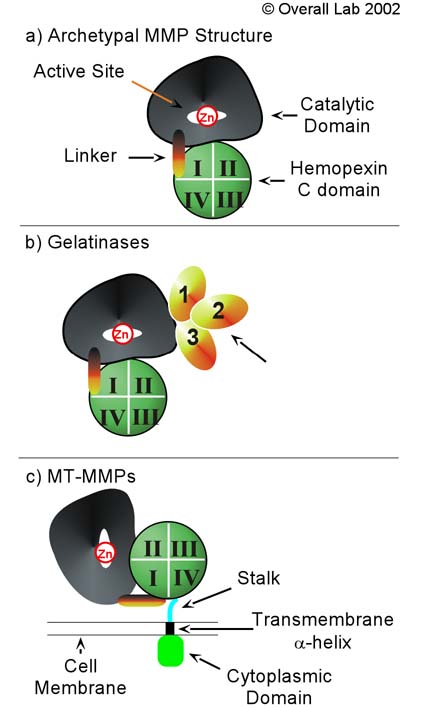
MMP's were first found in vertebrates in 1962 but have also been found in invertebrates and plants. They are distinguished from other similar enzymes by their dependence on metal ions as cofactors, their ability to degrade extracellular matrix, and their specific evolutionary DNA sequence. MMPs are typically secreted as inactive proproteins which are later activated when cleaved by another protease [3]. StructureThe structure of human MMP-1 was determined to have two monomeric structures (chains A and B). However, it is believed that this dimer is not physiologically relevant, as it was found that human MMP-1 is a monomer in solution. The structure of MMP-1, just like the other members of matrix metalloproteinases family, is formed by three different domains [3]. The structure consists of a , a variable and the . The catalytic domain of one monomer is right next to the hemopexin-like domain of the other monomer and is linked by the linker region. Other structural characteristics of MMP-1 include the many found on the protein. These structures were determined by using X-ray crystallography and NMR [3]. The basic structure of a MMP in three different forms/organisms.
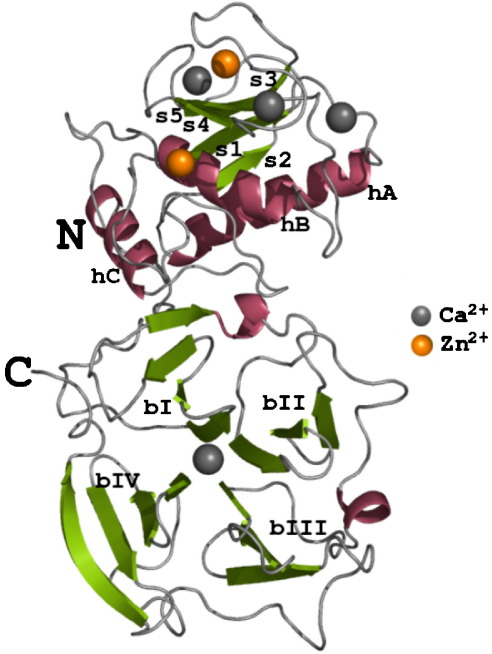
Catalytic Domain The Catalytic Domains of all MMPs share very similar characteristics, having the same general shape and a diameter of ~40Å. The of MMP-1 is composed of five highly twisted , three and a total of eight loops, enclosing a total of five metal ions, three Ca2+ and two Zn2+, one of which with catalytic role [4]. The Catalytic Domain (CAT) of MMP-1 starts with the F100 as the first amino-acid of the N-terminal loop of the CAT domain. This is different from the first published x-ray structure of the CAT domain which showed the shorter form of this domain, where the first 7 amino-acids are not present [5].
Taken from a publication of the crystal structure [4].
Linker region
Hemopexin-like domain The starts with Cys259 and forms a complete circle by joining to Cys447 using a disulfide bond that connects and gives this domain the characteristic four-bladed β-propeller structure. β-Propeller structures provide a large flat surface that is thought to be involved in protein-protein interactions [4]. This determines substrate specificity and is the site for interaction with TIMP’s (tissue inhibitor of metalloproteinases). Each blade starts with a , in which the Asp residues (Asp266, Asp359 and Asp408) direct the central calcium ion through their carbonyl oxygen atom. Glu310 provides the fourth coordination thus completing the acidic patch at the entrance of the central, solvent-accessible channel. The side-chains of these residues form salt bridges to the neighbouring β-strands holding the entrance of the central channel together. Three water molecules are found in the center of this channel but these molecules do not seem to be involved in this process. Two of the water molecules are at positions corresponding to the sodium and chloride ion in the proMMP-1 structure. The water molecule corresponding to the sodium ion is at hydrogen-bonding distances to the carbonyl oxygen atom of Ile268, Ala312, Ala361 and Val410 [4]. Mechanism of Action
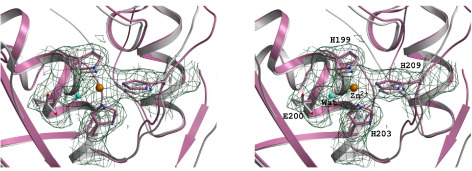
In the first mechanism, Browner M.F. and colleagues proposed the base-catalysis mechanism involving the conserved glutamate residue and the Zn2+ ion [2]. In the second mechanism, the Matthews-mechanism, Kester and Matthews suggested an interaction between a water molecule and the Zn2+ ion during the acid-base catalysis [2]. In the third mechanism, the Manzetti-mechanism, Manzetti Sergio and colleagues showed that the interaction between water and zinc during catalysis was unlikely. They then suggested a third mechanism where a histidine participates in catalysis by allowing the Zn2+ ion to assume a quasi-penta coordinated state. In this state, the Zn2+ ion is coordinated with the two oxygen atoms from the catalytic glutamic acid, the substrate's carbonyl oxygen atom, and the two histidine residues, and can polarize the glutamic acid's oxygen atom, proximate the scissile bond, and induce it to act as reversible electron donor. This forms an oxyanion transition state. At this stage, a water molecule acts on the dissociated scissile bond and completes the hydrolyzation of the substrate [2].
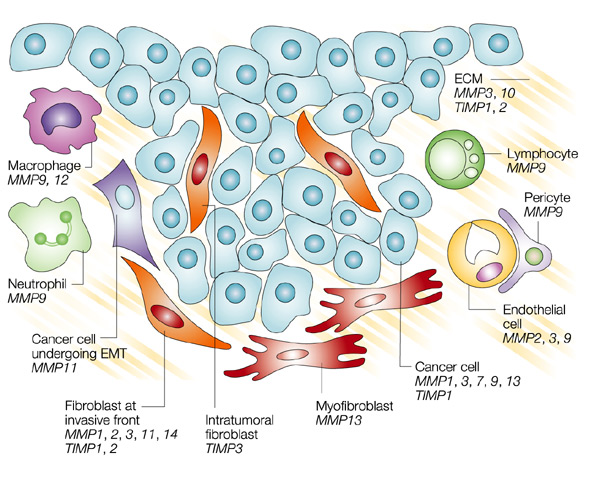
Inhibition MMPs are inhibited by molecules known as tissue inhibitor of metalloproteinases (TIMPs), which comprise a family of four protease inhibitors: TIMP-1, TIMP-2, TIMP-3, and TIMP-4. There are also synthetic inhibitors which generally work by binding the catalytic zinc atom at the MMP active site tightly and preventing catalysis. The common groups used for synthetic inhibition include hydroxamates, carboxylates, thiols, and phosphinyls. Hydroxymates are particularly potent inhibitors of MMPs and other zinc-dependent enzymes, due to their bidentate chelation of the zinc atom [2]. Medical ImplicationsResearch has shown that MMP-1 may have many medical implications. MMP-1 plays an important role in many physiologic processes such as development, tissue morphogenesis, wound repair and the remodeling of collagenous extracellular matrix. The enzyme is expressed by over 30 different cells [6]. Therefore, it is quite obvious that if this enzyme is not functioning properly then these processes will be affected, which could have little to no effect or have devastating effects. This figure shows the different cells that MMPs are expressed in.
MMP-1 gene expression has also been shown to have implications with cancer treatment. MMP-1 can be used as a candidate marker that may be useful for identification of breast lesions that can develop into cancer. As such, the medical implications of MMP-1 go beyond just what happens if the enzyme functions improperly but the enzyme, well rather the gene that encodes the enzyme, can also aid the diagnosis and subsequent treatment of breast cancer [2]. References
|