Sandbox Reserved 488
From Proteopedia
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
c-Myc ProteinBackgroundc-Myc protein, also known as Transcription factor p64, is a product of the c-myc proto-oncogene that has the ability to bind to DNA and regulate gene transcription.[1] More specifically, c-Myc recognizes the DNA's core sequence 5'-CAC[GA]TG-3' by forming a heterodimer with MAX, a bHLh protein. This structure then interact with TAF1C, SPAG9,PARP10, KDM5A, and KDM5B. This protein is found mainly in Human and it's known to express the MYC gene using an E.coli expression host. [2] Structure
The c-myc gene is consisted of three exons located on human chromosomes 8q24. The c-Myc sequence contains many Myc boxes, or conserved N-terminal domains, which are found in N-Myc and L-Myc.[3] The C-terminal region of c-Myc contains the helix-loop-helix, leucine zipper, and a basic region. These three components dimerize to form a bHLH LZ motif. The binding of DNA is achieved by the combination of the bHLH LZ motif and another bHLH LZ protein, Max through .DNA-bound Myc-Max complexes activate transcription through the amino terminal 143 amino acids of c-Myc, known as the transcriptional activation domain (TAD). [4]
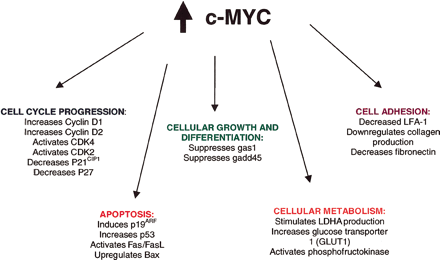
Molecular FunctionsOnce the Myc-Max complex binds to the specific DNA hexanucleotide core sequence, called the E boxes, the transcriptional activation domain then permits interaction with specific elements of the transcriptional machinery to allow transcription of nearby growth-related genes. c-Myc can also function as a transcriptional repressor of growth-inhibiting genes, not just an activator. [6] Expression of c-myc in the normal cell is regulated by external signals. [6] High levels of m-Myc activity might lead to proliferation and inhibition of differentiation while diminished levels of c-Myc activity lead to loss of proliferative capacity and to terminal differentiation. The c-Myc protein is involved in cellular transformation and mitogenesis, but is also a potent inducer of programmed cell death, or apoptosis.[7] C-Myc protein is also the expression of c-myc gene for normal embryonic development. Cancer DevelopmentThe c-Myc protein or the c-myc gene is overexpressed in a wide variety of human cancers with 80% of breast cancers, 70% of colon cancer, 90% of gynecological cancers, 50% of hepatocellular carcinomas and a variety of hematological tumors possessing abnormal myc expression.[4] The genesis of lymphoid malignancies is caused by translocations that juxtapose the c-myc proto-oncogene at chromosome 8q24 to one of three immunoglobulin genes on chromosome 2, 14, or 22 in B cells activate the c-myc gene. The c-myc gene is a central oncogenic switch for oncogenes and the tumor suppressor APC. The APC tumor suppressor protein mediates the degradation of β-catenin. The Wnt oncoprotein is shown activating its receptor, which results in the stabilization of free β-catenin. β-Catenin, which sustains activating mutations in human cancers, is a cofactor for the transcription factor Tcf. Tcf activates c-myc expression through specific DNA binding sites. [3] In addition to activation of the c-myc gene through deregulated expression, point mutations in the coding sequence have been found in translocated alleles of c-myc in Burkitt’s lymphomas. [8] References
|