Sandbox Reserved 499
From Proteopedia
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
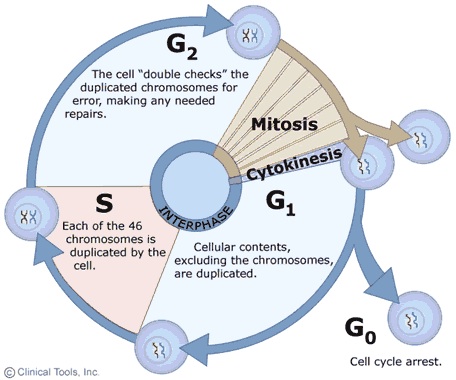
Cyclin-Dependent Kinase 3INTRODUCTIONCyclin-dependent kinase (Cdk) is a protein kinase family involved in regulation of the cell cycle. These Cdks are relatively small protein enzymes are responsible for mitosis, specifically transcriptional activity. They help activate DNA repair by being inhibited through phosphorylation during DNA damage response (DDR). They are also generally inactive during checkpoints along the cell cycle. More than one Cdk can be in charge of the different phases[1]. Often times, DDR is controlled by overall level of Cdk activity, and not by individual Cdks. Cdk’s are found in Eukaryotes, including human and yeast cells. Each organism has a different number of Cdks. In yeast, there is less, and more in humans. It is easier to study which Cdk is responsible for which part of the cell cycle in yeast than in humans, because of the level of complexity of Cdk and other cell activities in Homo Sapiens [1]. However, the function of Cdk is similar in all organisms in that it is part of the cell cycle regulation. For this reason, Cdk is often studied as targets for cancer treatment.
MECHANISM/ PATHWAYCDKs are activated through physical association with cyclin, a family of proteins, in which their abundance and proteolysis pattern within the cell vary according to the stage of cell cycle. Major transitions in the cell cycle are caused by activation of CDKs 1-4, which causes phosphorylation and dephosphorylation of key residues. Cells enter mitosis as protein-bound phosphates increase[3]. In mammalian cells, it had been suggested that CDK-3 works alongside CDK-2[4], which is in charge of the G1/S stage to regulate transcription factors[5]. It had been shown that it delays cells in the G1 phase[6]. It is partnered with CDK3-Cyclin C to control G0/G1 transition, or reentry to the cell cycle. The CDK3-Cyclin C complex phosphorylates protein targets such as pRB (retinoblastoma protein) that lead to the release of transcription factor E2F [5].
STRUCTURECurrently, no crystal structure for human CDK-3 had been resolved. There is only a theoretical molecular model available, based on analysis of 39 binary complexes of CDK-3:inhibitors. Data showed that 74.18% of CDK-3 structure is sequentially identical with CDK-2, thus making CDK-2 a perfect template model for CDK-3 protein [5]. The Parmodel web server was used for comparative modeling and evaluation of protein structure, and molecular dynamics (5 ns) simulation was done with GROMACS[5]. Based on the molecular model, the overall structure of human CDK-3 was found to contain 305 amino acid residues, with a total molecular weight of 35,045.74 Da, and a theoretical pI of 8.86. Many features of secondary structure and molecular fork was found to very much resemble CDK-2[5].
Human CDK-3 proteins was found to have alpha and beta structures. Its fold contains two alpha and two beta domains, with a that is mostly alpha helical (this is characteristic of a typical protein kinase fold). CDK-3 is found to be folded into a bilobal structure, with smaller N-terminal lobe that is mostly β-sheet structure[5]. The is found in a sheet of 5 antiparallel β-strands (β1-β5), and in one, large alpha helix (α-1). The C-terminal domain contains pseudo-4-helical bundle (α-2 to α-3, α-6), small β-ribbon (β-6 to β-8), and 2 alpha helices (α-5, α-7)[5]. BINDINGATP is a cofactor for CDK3. It forms 17 intermolecular bonds with the ATP binding pocket is found in the cleft in between the bilobal structure. Most residues found in the ATP binding pocket is hydrophobic. The hydrophobic pocket can fit in different geometries of residues, like adenine derivatives and flavonoids. It is determined so far that in the binding of ATP to CDK3, an are present[5]. Binding between CDK3 and inhibitors at the molecular fork also require intermolecular hydrogen bonds. They are responsible for specificity and affinity between the protein and its ligand. Although specific contact areas have not yet been concluded, a high correlation coefficient between CDK-3 and CDK-2 pkd values suggest that CDK-2 inhibitors can also inhibit CDK-3. Pkd values for various known CDK-2 inhibitors with were analyzed in its interaction with CDK-3, and 4 inhibitors were shown to be the best[5]: - n-methyl-{4-[2-(7-oxo-6, 7-dihydro-8h-[1,3]thiazolo[5,4-e] indol-8-ylidene) hydrazine]phenyl}methanesulfonamide - (2r)-1-(dimethylamino)-3-{4-[6-{[2-fluoro-5-(trifluoromethyl)phenyl] amino}pyrimidin-4-yl)amino]phenoxy}propan-2-ol - (5-chloropyrazolo[1,5-a]pyrimidin-7-yl)-(4-methanesulfonylphenyl)amine - din-232306 6-(3,4-dihydroxybenzyl)-3-ethyl-1-(2,4,6-trichlorophenyl-1h-pyrazolo[3,4-d]pyrimidin-4(5h)-one
MOLECULAR DYNAMICS
Molecular dynamics of human CDK-3 protein model was assessed by RMSF values. Backbone fluctuations, or residues of higher flexibility, occur in loop and turn regions of residues that surround the alpha-beta-alpha fold. Relatively high RMSF values are found in more flexible regions in the CDK-3 structure. These regions are: L1 (turn composed of Glu25- Gly27), L2 (loop Leu37- Pro45), L3 (turn Glu73- Arg74), L4 (loop Thr94- Pro100), L5 (turn Tyr159- His161), and L6 (loop Gly220- Gly247)[5]. MEDICAL IMPLICATIONBecause of its significant role in the regulation of cell cycle, CDK-3 is often a target for cancer research. Protein expression level of CDK3 is shown to be high in human cancer cell lines and human glioblastoma tissue[8]. It is currently a target for many cancer therapies, including for multiple myeloma (MM). Study had shown that a multi-targeting CDK inhibitor, LCQ195, can inhibit CDK3, along with CDK1, CDK2, CDK5, and CDK9, leading to cell cycle arrest and apoptotic MM cell death[9]. There are currently high hopes for curing various types of cancer based on targeting CDK3, and its other family members. REFERENCES
|