Sandbox Reserved 500
From Proteopedia
| This Sandbox is Reserved from 13/03/2012, through 01/06/2012 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 451 through Sandbox Reserved 500. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://www.proteopedia.org/wiki/index.php/Help:Getting_Started_in_Proteopedia
Protein 53 (p53)IntroductionCancer is one of the most popular diseases occurring today. Research has been done on this issue for an extended amount of years to figure out how this devastating case works and what factors are contributing to this heartbreaking illness. Throughout time there have been some successful answers while some things still remain in question during this existent investigation. One of the most interesting roles in the fight with cancer happens to be a protein quite phenomenal to cell life. This protein is none other than the unique protein 53 or p53. Protein 53 is known for its function as the tumor suppressor gene which helps to prevent cancer. P53 protein is an extremely important system in cell cycle control and the area of cancer. This protein was researched by Bert Vogelstein, David Lane, and Arnold Levine.
Guardian of the GenomeProtein 53 is a DNA binding protein that helps to prevent cancer. It is often called the guardian of the genome because of the role it has in checkpoint pathways of the cell cycle. This protein is located on chromosome 17 on the short arm which is where the open reading frames occur for proteins. The p53 protein was discovered through co-purification with a large T antigen in SV40 virus transformed cells. Protein 53 earned its name from running on the SDS-PAGE with a molecular mass of 53 kilodaltons. However it was found that the mass of p53 is less because of the amount of proline amino acid residues it has which as a result slow down its migration on the gel. P53 protein is central for multicellular organisms such as humans, cows, fish, and rodents.
StructureThe structure of the p53 protein is pretty interesting. This protein has a single polypeptide chain divided into three discrete domains, which show a structure that has helped to facilitate study in the absence of the rest of the protein. These domains have been found to be a concern with tetramerization, transcriptional activation, and DNA binding. In the there are two helices and ten beta strands making up two beta sheets. In the p53 protein an acidic N-terminal transactivation domain extends from residues 1-99. When broken down the N-terminal has two complementary transactivation domains, a major and a minor, which are involved in the regulation of the pro-apoptotic genes. This is then followed by the central core region, also referred to as the central DNA-binding core region, which is the largest domain. This area contains residues 100-300 that bind specific DNA sequences. The central core region is responsible for binding the p53 co-repressor. The third region of this protein is the C-terminal domain from residues 301-393, which includes a regulatory region and a tetramerization domain. The C-terminal domain helps facilitate downregulation of DNA binding that occurs in the central domain. These are the main three domains described for this protein, it has been disputed that there are actually four up to seven domains instead of three. Discrete domains are linked by flexible linkers creating a molecule with dynamic conformation. The p53 protein has been found to be difficult to crystallize when in the form of its full length. The p53 protein is a β sandwich formed by the interaction of antiparallel four and five stranded elements of the β sheet. The essential part of this protein is the where the ligands are clustered together. There are three particular parts of the active site, starting with the which is bound to at least four water molecules but then also contains the leucine, glutamine, and glycine amino acids. The consists of the lysine, leucine, arginine, glutamine, tyrosine, and tryptophan amino acids. The contains one water molecule, two lysine residues along with serine, glutamine, and arginine amino acids.
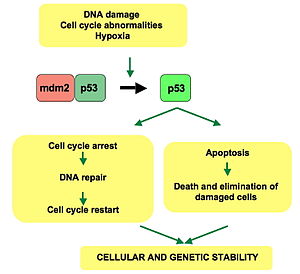
FunctionThe p53 protein is a tumor suppressor gene that stops the formation of tumors, particularly tumors that can lead to cancer. It becomes activated in response to stress which includes DNA damage. In order for the p53 protein to work it has to be phosphorylated. When DNA has double-stranded breaks, its altered form triggers the activation of the ataxia telangiectasia mutated (ATM) protein kinase. The ATM enzyme catalyzes the phosphorylation of checkpoint kinases within the cycle, which as a result phosphorylate the p53 protein. Phosphorylating the p53 protein in this manner helps protect it from degradation allowing it to buildup in the presence of damaged deoxyribonucleic acid (DNA). Now that p53 is available it can carry out its task in the cell cycle. The function of the p53 protein once it has accumulated includes cell cycle arrest in the G1 phase of the cycle and cell death. These events involve p53 binding to DNA and acting as a transcription factor which stimulates transcription of specific genes. In cell cycle arrest, the p53 protein binds to DNA activating the transcription of the gene that codes for a Cdk inhibitor known as p21. The p53 protein causes inhibition of the p21 protein preventing phosphorylation of the Rb protein. This results in cell cycle arrest at the restriction point. The other event which involves the p53 protein is cell death. This process occurs when the damage DNA is unable to be repaired of an error. As a result the p53 protein activates gene coding for a group of proteins which use apoptosis to trigger cell death. However, p53 does have an aid in completing this event successfully by working along with the p53 upregulated modulator of apoptosis (called Puma for short), which binds to and inactivates the Bcl-2 protein and promotes apoptosis. In order for any of these processes to occur it is very important that p53 is phosphorylated to avoid degradation. Phosphorylating the p53 protein will prevent it from interacting with the Mdm2 protein, which if allowed will link p53 to ubiquitin marking it for destruction by the proteasome. Having as much p53 protein as possible is not necessarily helpful. Trying to increase the amount of p53 present may seem like a good way to treat tumors or prevent them from spreading, but in actuality it is not a usable method for treatment. Having too much p53 protein can cause premature aging; however it does help to restore the function of the protein. Loss of p53 can create genomic instability. ApplicationsP53 protein has been used in other experiments to help find cures for cancer and identify certain genes which are based on if the p53 protein is expressed. The most popular research done using this protein is of course cancer research. The p53 protein has been used as a biomarker for types of cancer such as bladder cancer. This protein makes great use for studying different types of cancer, however there have been some difficulties that occurred in the process. In the bladder cancer research where p53 was used as a biomarker there were different issues which included the evaluation of markers and the measurement of p53 because of differences in assay, study design, and analysis. As a biomarker, p53 was found to be difficult to measure because of its diverse mutations and doing certain staining does not always identify all mutations. Mutations in the p53 protein tend to occur in pathogenesis of the transitional cell carcinoma for the bladder which could help in spotting cancer development. Overall the main use for the p53 protein is for anticancer research. Researchers have performed experiments over the years to determine how mutations in the protein affect the rest of the processes of the cell cycle and if there were ways to improve the protein to make it more efficient in the battle against cancer.
References
|