Sandbox Reserved 592
From Proteopedia
==Your Heading Here (maybe something like 'Structure'-- PLEASE DO NOT DELETE THIS TEMPLATE -->
| This Sandbox is Reserved from Feb 1, 2013, through May 10, 2013 for use in the course "Biochemistry" taught by Irma Santoro at the Reinhardt University. This reservation includes Sandbox Reserved 591 through Sandbox Reserved 599. |
To get started:
More help: Help:Editing |
Contents |
Background
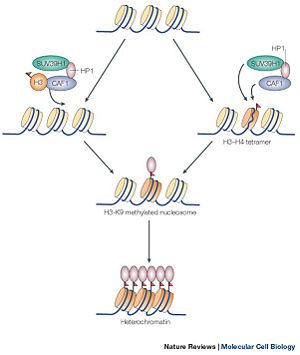
SUV39h1 is part of a class of methytransferase that deals with the methylation of histone proteins in nucleosomes. It is one of the very first histone methyltransferase to be discovered [1]. The methylation of histone proteins is an essential part of an area of genetics which deals with the modification of histone proteins called epigenomics. Epigenomics is the study of modifications of nucleosomes, which either inhibit or express gene transcription without changing the underlining DNA sequence. These changes are allowed because of the amino acid tails which extend out and away from the histone proteins, exposing itself to methylation. Methylation of histone protein is one of many ways that regulates gene expression [2]. The regulation of gene expression is dependent on the state of the gene. A gene cannot be transcribed when the promoter of the gene is wrapped around the nucleosome, forming a heterochromatin state; thus, the gene is inhibited from being transcribed. Moreover, Methylation of the histone protein causes the winding around the nucleosomes, transforming a euchromatin state into a heterochromatin state. Of course, SUV39H1 alone does not methylate the histone proteins. SUV39H1 interacts with other proteins in order to efficiently methylate a histone protein (shown in Figure 1). Abnormal expression of genes, which is the result of down regulation or mutation of SUV39H1, results in a variety of diseases [3].
Structure
SET Structure
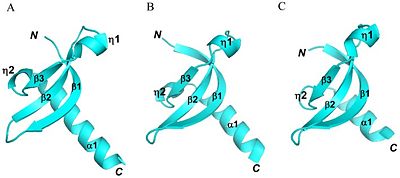
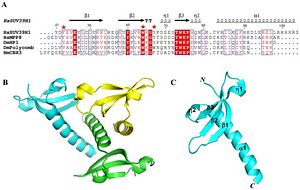
There is a chromodomain and SET domain. The SET is known as the catalytic domain. The catalytic domain is consisted of an alpha double helix. The double helix is located at the C-terminus end of the protein. The residue consisting the catalytic domain is about 82-100 amino acids (Shown in Figure 2). The C-terminus is where the transfer of the methyl group to the lysine residue occurs. In addition to the two main domains to the enzyme, there is an essential hydrophobic core which is very similar to other chromodomain proteins. The hydrophobic core is made up several residues. These residues are V45, L48, Y60, V62, W64, L80, I85 and L86 (Shown in Figure 2) [4]. The groove formed by the beta sheets is also similar and conserved feature of the chromodomain family. The physical characteristics between the chromodomain of SUV39h1 and chromodomains of other enzymes are very similar. SUV39h1 has been shown to very similar to the chromodomain of MPP8 and HP1, showing a conservation in chromodomain structure (shown in Figure 3). Although the chromodomain structure is very similar, there is a slight difference with the catalytic domain being longer. In addition to the catalytic domain of SUV39H1 being longer, the enzyme does not contain a F34 aromatic cage, which was originally thought to be essential for recognizing exposed lysine or argon residue. However, crystallography of residues (44-106) has shown residues missing the F34 aromatic cage. Instead, it has W64 and Y67 which form a loop ; this is also another conserved feature among chromodomain family [5].
|
Chromodomain Structure
Crystal structure shows two main parts to the protein, but as a whole three independent tertiary protein molecules; the total length of the protein is 44-106 amino acids [6]. The chromodomain starts at the N-terminus of the enzyme and continues toward the C-terminus, where the SET catalytic domain is located. The chromodomain length is around 44-80 amino acids, which forms three anti- parallel beta sheets. The residues for each of three beta sheets are 45-53, 58-64 and 73-76 amino acids for beta 1, beta 2 and beta 3, respectively ( shown in figure 2). These three beta sheets form the chromodomain.
|
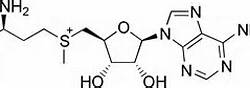
FunctionSET FunctionThere are two types of histone methyltransferases: lysine specific and argine specific; each is a residue which the enzyme transfers a methyl group to. Within the lysine specific, there are further two main types: SET (Su(var)3-9, Enhancer of Zeste, Trithorax) and non-SET domains. These domains are an essential part of the enzyme because the main catalytic occurs in SET domain of the methyltransferase. SUV39h1 is considered to have a lysine specific SET domain which catalyzes the methyltransferase. In the nucleosomes, there are four different proteins; each consists of two copies which make the entire nucleosome. The mechanism by which SUV39H1 methylates the histone proteins is also known. A nearby tyrosine residue in the histone protein creates a strong nucleophile by deprotonating a nearby lysine residue in the same histone protein. The lysine residue is a very strong nucleophile which attacks the sulfur atom of the cofactor S-Adensyl methoinine (SAM) and extracts a methyl group from the cofactor. Subsequently, the attack methylates the histone protein (Shown in Figure 4). The cofactor is very important because it is the source of the methyl group.[7] Chromodomain FunctionThe chromodomain structure of the enzyme (residues) is not involve in the catalytic function of the enzyme, but is very important to the binding to the molecule. Mutations which remove the chromodomain area of the enzyme result in the inhibition of the enzyme1 [8]. The inhibition of the chromodomain prevents the methylating action of the enzyme's catalytic domain. Although the chromodomain is not directly involved in the methylation of the histone proteins, it plays an important role in the binding of the enzyme to histone proteins as well as other proteins. Fluorescence Polarization Assays have shown the chromodomain has a high affinity for the histone protein H3K9me1 and H3K9me2. SUV39H1 residues that did not contain the aromatic F34 cage did not bind to the histone protein efficiently. This shows the F34 is a conserved residue that is essential for the enzyme binding to the histone protein. Moreover, a mutation which results in the loss of the aromatic F34 cage group will inhibit the binding effect of the family of chromodomain enzymes. SUV39H1 does not methylate the histone protein alone; it is aided by other proteins [9]. Studies have shown HP1 and H3 are essential proteins that combine with SUV39H1 in protein complex, which methylate the histone protein of a nucleosome [10].
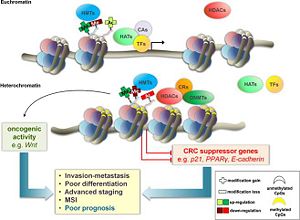
Clinical RelevanceThe physiological state of a cell depends on the physiological state of the genes which defines the function of the cell. Unregulated expression of genes in the cell leads to physiological and morphological change of the cell. Euchromatin and heterochromatin define the physiological state of a cell. Mutations and knockout studies of SUV39H1 in mice have shown to increase genomic instability. In humans, mutations importantly result in tumorgenesis. The instability arises from the inability of SUV39H1 to form heterochromatin. Since the enzyme could not methylate the nucleosomes and transform a euchromatin state into a heterochromatin state, certain genes were highly expressed, which lead to the instability of cells [11]. Further knockout studies of SUV39H1 have shown that length of telomeres in mice and pigs have increased. The increase in the length of the telomeres also increases tumor genesis in pigs and mice. The previous studies, however, did not show a significant telomere growth in human cells. The results may indicate an epigenetic regulation that is more similar between pigs and mice than human [12]. The results from previous studies in knockout mice further support cancer related symptoms. In colorectal cancer (CLC), Unregulated SUV39H1 lead to the gene silencing of tumor suppressor genes of CLC. The silencing of the tumor suppressor gene leads to the proliferation and subsequent promotion of CLC (Shown in Figure 5)[13]. Chaetocin was shown to inhibit SUV39H1 from binding to H3K9. There is also siRNA and other gene therapies which showed inhibition of SUV39H1 [14][15]. Morover, numerous compounds have been synthesized or discovered which inhibit numerous parts of histone methyltransferases. BIX01294 is a compound that blocks protein-protein interaction. It does not inhibit suv39h1 as well as other methyltransferases, but inhibiting other methytransferases can inhibit the transfer of methyl groups, since histone methyltransferases require other proteins[16]. Also, replacing the essential cofactor SAM would effenciently inhibit the histone methyltransferases. EPZ004777 replaces the SAM in a methytransferase that is not SUV39H1 (DOT1L) ; however, DOT1L binds with other proteins (SUV39h1) to methylate the nucleosomes [17]. References
|