Sandbox Reserved 660
From Proteopedia
| This Sandbox is Reserved from 30/08/2012, through 01/02/2013 for use in the course "Proteins and Molecular Mechanisms" taught by Robert B. Rose at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 636 through Sandbox Reserved 685. | |||||||
To get started:
More help: Help:Editing For more help, look at this link: http://proteopedia.org/w/Help:Getting_Started_in_Proteopedia
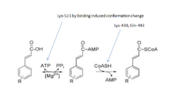
4-coumarate:CoA ligase4-coumaroyl-CoA (4CL) ligase belongs to the family of ligase and is an enzyme that catalyze the reaction below: ATP+4-coumarate+CoA <--> AMP+ diphosphate+4-coumaroyl-CoA. Ligase in the enzymology, is an enzyme that catalyze the joining of two large molecule by forming new chemical bond. In the process of catalysis, there is always hydrolysis of small molecule group accompanying. 4CL ligase, sepcifically, catalyzes the formation of carbon sulfur bond between two reactants. 4CL is one of the key enzyme regulating the synthesis of monolignols in the lignin pathway. Lignin is an unique and complex phenylpropanoid polymer which plays key role in plant development and response to the environment. Lignin is typically polymerized from 3 phenylpropanoid monomers , p-coumaryl (H), coniferyl (G) and sinapyl (S)[1]. The relative amounts of these 3 monomers found in lignin show plant specificity and 4CL is one of the main enzymes participating the formation these 3 monomers in the regulatory network of the monolignol pathway and has significant effect on the S/G ratio which is substantial to the lignin formation in the plant. There is report showing that the 4CL gene knock-down will result in the lignin reduction[2]. In other words, the activity of 4CL determines the overall carbon flow of the phenylpropanoid pathway. For this reason, 4CL has been focused on the genetic engineering regulation about the plants products quality improvement.
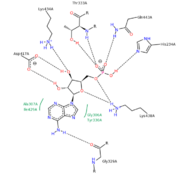
StructureThe P. tomentosa 4CL1 protein consists of 536 residues and the overall structure of 4CL1 consists of two distinctive globular domains, the large N-domain and the small C-domain. The N-domain contain 3 N1, N2 and N3. N1 and N2 are the structural similarly larger subdomains and the N3 is the smaller one. Each of these two subdomains has a central eight-stranded (6 parallel and 2 anti-parallel β -strands) that is flanked by two on one side and four α-helices on the other. Subdomain N3 is mainly consisted of β -barrel formed by eight β-strands and an α-helices and 310 helices are also found in this subdomain. Center of the C domain of 4CL1 is a 3-stranded mix β-sheet with a combine of 2 α-helices and 2 antiparallel β–sheet on one side of the stranded mix β–sheet and a single α-helices on the other side[3]. AMP-binding pocketThe AMP is mainly located within the N-domain. The highly conserved sequence which corresponds to in 4CL provides the majority of the surface of the binding pocket. The side chain phenyl group of Tyr-330 provide the Van Der Waals interaction to the adenine group of the bound AMP on one side of the adenine ring and on the other side, the adenine group is support by the side chain of Gly-306 and main chain of Ala-307. The N atoms of adenine group could also donate hydrogen bond to the carbonyl oxygen of Gly-329. Several hydrogen bonds are formed between the ribose oxygen atoms and the side chains of Arg-432, Lys-434, and Lys-438 and the phosphate group could also form hydrogen bond with side chain of Thr-333 and Gln-443 and main chain NH of Thr-333[3]. The of the 4CL is located on Lys-523 within the C domain and Lys-438 and Gln-443 within the N domain. there is a Ligand Binding–Induced Conformational Changes observed in 4CL1 structures upon the binding of AMP. A large cleft was observed between the N- and C-domains in apo-4CL1 (unmodified) and the cleft is closed by an 81˚ rotation of the C-domain relative to the N-domain when the ligand AMP is bound to the binding pocket. Because no major internal conformation or structure changes are found in either the N- or C-domain, the closure of the inter domain cleft upon the binding of AMP is a rigid-body movement[4]. The Inter domain movement brings different catalytic residues (i.e. Lys-523) from the C-domains to substrate binding sites to catalyze the respective partial reactions, such as the adenylate-forming partial reaction, and the thioester-forming partial reaction.
Enzymatic MechanismThe enzymaticmechanism begins with the binding of ATP and hydroxycinnamate substrates. The binding results in 4CL1 adopting the catalytic conformation for the adenylate-forming partial reaction, in which the side chain of Lys-523 interacts with and directs the carboxylate group of the bound hydroxycinnamates for the nucleophilic attack of the α-phosphate of ATP, resulting in an AMP-hydroxycinnamate conjugate and a PPi molecule. The release of PPi then propels 4CL1 to the catalytic conformation of the thioester-forming partial reaction. In this conformation, the side chain of His-234 swings aside to allow access of CoA to the AMP-hydroxycinnamate conjugate. The AMP-hydroxycinnamate conjugate and CoA are then catalyzed by side chains of Lys-438 and Gln-443 to form the final thioester product. The C-domain rotates again to expose the substrate binding site, and the thioester and AMP are released[3]. Implication4-Coumarate-coenzyme A ligase (4CL) is an enzyme that functions early in the general phenylpropanoid pathway by producing the monolignol precursor p-coumaroyl-CoA. 4CL silencing in angiosperm species such as tobacco, Arabidopsis and Populus tremuloides causes lignin reductions in the range of 25% to 45%. However, the impacts of these manipulations on lignin composition varied. 4CL silencing in tobacco preferentially depleted sinapyl (S) lignin units, which are prominent in wood fibers. 4CL silencing in Arabidopsis depleted only coniferyl (G) lignin units, which are enriched in vessel elements, whereas silencing of 4CL in P.tremuloides had no impact on the S-G ratio. An increase in the S-G ratio in P.tremuloides was only recorded when 4CL silencing was combined with the over expression of coniferaldehyde 5-hydroxylase. These differences in lignin composition could be the consequence of silencing 4CL isoforms with different substrate preferences, or they could reflect the in adequacies or limitations of analytical procedures used for lignin analysis[5].
references1. Ralph, J., Brunow, G., Harris, P. J., Dixon, R. A., Schatz, P. F., and Boerjan, W. 2008. Lignification: Are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? In F. Daayf, A. El Hadrami, L. Adam and G. M. and Ballance (eds.), Recent Advances in Polyphenol Research. Wiley-Blackwell Publishing, Oxford, UK. 2. Ralph, J., Lundquist, K., Brunow, G., Lu, F., Kim, H., Schatz, P.F., Marita, J.M., Hatfield, R., Ralph, S.A., Christensen, J.H., and Boerjan, W. 2004. Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3:29-60. <http://www.fpl.fs.fed.us/documnts/pdf2004/fpl_2004_ralph001.pdf > 3. HU, Y.; GAI, Y.; YIN, L.; WANG, X.; FENG, C.; FENG, L.; LI, D.; JIANG, X.N. and WANG, D.C. (2010). Crystal structures of a Populus tomentosa 4-coumarate:CoA ligase shed light on its enzymatic mechanisms. The Plant Cell, vol. 22, no. 9, p. 3093-3104 < http://www.ncbi.nlm.nih.gov/pubmed/20841425 > 4. Gulick, A.M. (2009). Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem. Biol. 4: 811–827 < http://pubs.acs.org/doi/abs/10.1021/cb900156h > 5. Wagner, A., Donaldson, L., Kim, H., Flint, H., Phillips, L., Steward, D., Torr, K., Koch, G., Schmitt, U., & Ralph, J. (2009). Suppression of 4-coumarate-CoA ligase in the coniferous gymnosperm Pinus radiata. Plant Physiology, 149, 370-383. <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2613735/> 6. http://www.rcsb.org/pdb/explore/explore.do?structureId=3a9v |