Sandbox Reserved 697
From Proteopedia
| This Sandbox is Reserved from 30/01/2013, through 30/12/2013 for use in the course "Biochemistry II" taught by Hannah Tims at the Messiah College. This reservation includes Sandbox Reserved 686 through Sandbox Reserved 700. |
To get started:
More help: Help:Editing |
|
Contents |
Monoamine Oxidases
Monoamine oxidases (MAOs) are a class of enzyme that are localized to the outer membrane of the mitochondria and catalyzes the oxidative deamination of different monoamines. Monoamine oxidase (MAO) plays a distinct role in depression, along with other psychiatric and neurological diseases. Inhibition of this enzyme increases levels of neurotransmitters in the central nervous system, such as serotonin, dopamine and norepinephrine. Monoamine oxidase inhibitors (MAOIs) have been used for decades as an effective treatment for depression. However, they are used somewhat infrequently due to safety concerns or dietary restrictions. More effective inhibitors of MAO are currently being researched. There are two subtypes of MAOs: and , which are closely related in structure, but each has different substrate and inhibitor specificities. The two subtypes of MAO shown were those of humans, which can vary between different species of mammals.
Structure and Function of MAO-As
MAO-A is a monomer that is made up of 506 amino acids total. Substrates for MAO-A include dopamine, serotonin and norepinephrine. The is located in the center of the protein shown in green. There are multitude of residues that create the of MAO-A and interact with the substrate. Among these residues is a cysteine, which has been shown to be especially vital for catalytic activity. The binds in this area, near where the flavin adenine dinucleotide (FAD) is bound. However, the molecule depicted is actually the reversible inhibitor harmine, but the substrate binds at this same site. The FAD binds at an shown in cyan which is crucial for the activity of the enzyme. The inhibitor can either be reversible and interact as harmine does, or it can bind covalently to FAD and be an irreversible inhibitor. MAOs are anchored into the outer membrane of the mitochondria by a shown in yellow. All of these domains are mostly conserved among MAO-As of different species.
Structure and Function of MAO-Bs
MAO-B is a dimer composed of 520 amino acids in each subunit. MAO-B also binds similar monoamines to MAO-A, but preferentially binds beta-phenylethylamines and benzylamine. The is shown in light blue, with the green molecule showing the location of where the substrate would bind. There are specific residues in the that interact with the substrate, allowing catalytic activity. Again, there are three cysteines necessary for enzymatic function, although two of these three are not in the active site. The is shown in green with the domain predicted to form the shown in red. The shown in yellow contains the FAD, which is covalently bound to either the substrate or inhibitor. The located in the center of the protein binds the inhibitor pargyline as shown. Pargyline is an irreversible inhibitor since it covalently binds to the FAD. These regions all play a vital role in the enzyme and are for the most part conserved.
Determination of Specificity between MAO-A and MAO-B
There is one crucial residue that is different between MAO-A and MAO-B that plays a crucial role in the specificity of each enzyme. In MAO-A, there is an in the 335 position, while in MAO-B, there is a in the 326 position that both determine the specificity. For example, the inhibitor harmine of MAO-A cannot be incorporated into MAO-B due to its structural overlap with the Tyr-326 of MAO-B.
Treatment for Depression and Other Neurological Disorders
MAO-A is primarily treated for anxiety and depression while MAO-B is primarily treated for Alzheimer's disease and Parkinson's disease. Some common inhibitors of MAO-A include clorgyline and moclobemide, while some inhibitors of MAO-B include selegiline and rasalgiline. Selegiline and moclobemide are some of the few used as drugs to help with neurological disorder. For the most part, drugs should be reversible inhibitors and selective towards the type of MAO they bind, so they allow some monoamine oxidase activity.
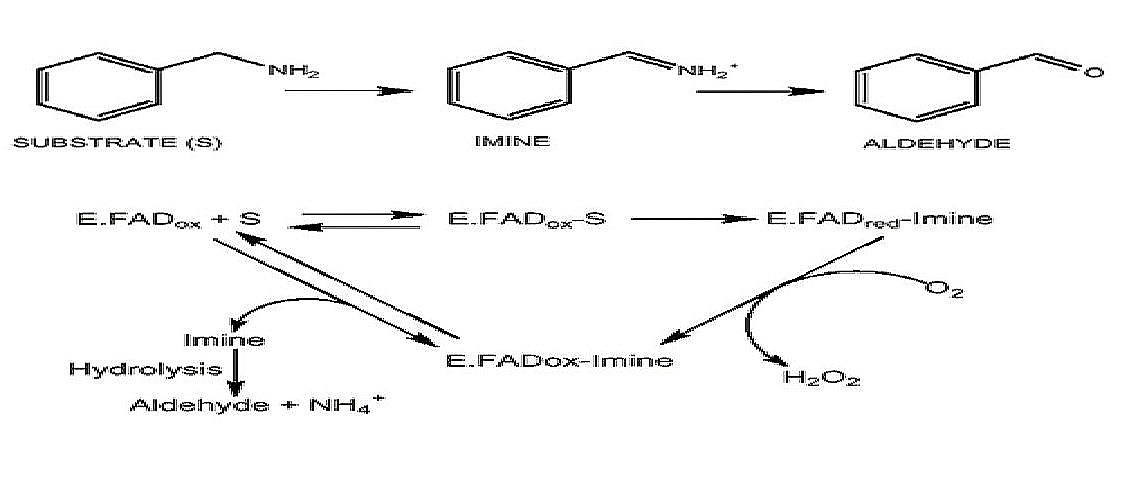
Mechanism of Action of MAOs
The top portion illustrates how a substrate for an MAO(in this case benzylamine) is converted to an aldehyde. This is an oxidative deamination reaction. The bottom part shows how the FAD portion of the active site in MAO is involved in the reaction to eventually release the aldehyde form of the monoamine. The FAD utilizes a sulfur from a nearby cysteine residue.(Figure adapted from Meyer 2006)
References
1. Binda, C., M. Li, F. Hubalek, N. Restelli, D.E. Edmondson, and A. Mattevi. Insights into the mode of inhibition of human mitochondrial monoamine oxidase B from high-resolution crystal structures. 2003. Proc Natl Acad Sci 100(17): 9750-9755.
2. Binda, C., P. Newton-Vinson, F. Hubálek, D.E. Edmondson, and A. Mattevi. Structure of human monoamine oxidase B, a drug target for the treatment of neurological disorders. Nature Structural Biology (9): 22-26.
3. Colibus, L.D., M. Li, C. Binda, A. Lustig, D.E. Edmondson, A. Mattevi. Three dimensional structure of human monoamine oxidase A (MAO-A): relation to the structures of rat MAO-A and human MAO-B. 2005. PNAS 102(36): 12684-12689.
4. Meyer, J.H., N. Ginovart, A. Boovariwala, S. Sagrati, D. Hussey, A. Garcia, T. Young, N. Praschak-Rieder, A.A. Wilson, and S. Houle. Elevated monoamine oxidase A levels in the brain: an explanation for the monoamine imbalance of major depression. 2006. Arch Gen Psychiatry (11): 1209-1216.
5. Son, S., J. Ma, Y. Kondou, M. Yoshimura, E. Yamashita, and T. Tsukihara. Structure of human monoamine oxidase A at 2.2-Å resolution: the control of opening the entry for substrates/inhibitors. 2007. PNAS (105): 5739-5744.
6. Walker W.H., E.B. Kearney, R.L. Seng, T.P. Singer. The covalently bound flavin of hepatic monoamine oxidase. 2. Identification and properties of cysteinyl riboflavin. 1971. Eur J Biochem 24:328–331.