Sandbox Reserved 786
From Proteopedia
| This Sandbox is Reserved from Oct 10, 2013, through May 20, 2014 for use in the course "CHEM 410 Biochemistry 1 and 2" taught by Hanna Tims at the Messiah College. This reservation includes Sandbox Reserved 780 through Sandbox Reserved 807. |
To get started:
More help: Help:Editing |
|
Contents |
Overview of Mitochondrial Malate Dehydrogenase
Malate dehydrogenase is an enzyme in the citric acid cycle. Overall it is classified as EC 1.1.1.37. Breaking down the classification shows precisely what the enzyme is responsible for doing. "EC 1" generally terms the enzyme as an oxidoreductase, a class of 7,911 protein database entries. An oxidoreductase is an enzyme that is responsible for catalyzing oxidoreduction actions. "EC 1.1" narrows the classification of malate dehydrogenase to an oxidoreductase that is responsible for acting on the CH-OH groups of substrates as donors, a collection of 1,754 protein database entries. "EC 1.1.1" furthers the specificity of the protein to be an oxidoreductase that acts on CH-OH groups using NAD(+) or NADP(+) as acceptors, a group of 1,585 entries in the protein database. The final classification"EC 1.1.1. 37" yields 60 protein database entries, and narrows the definition with the name of the enzyme,malate dehydrogenase, and its specific reaction.
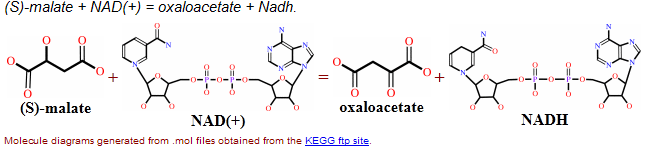
Enzymatic Reaction
Overview of Crystalline Structure
The shows that it is a homo-tetramer. However the is a dimer. In the image, the alpha-helices are green, Beta-sheets are red, and random coils (turns) are grey.
Secondary Structure
The is formed by hydrogen bonding interactions of the amino acids backbone. There are two beta-sheet regions in each monomer of the natural dimer. Five beta-sheets in each monomer are anti-parallel, and form a pseudo beta-barrel motif. Six beta-sheets in each monomer show parallel configuration. The angle of the hydrogen bonds relative to one another between beta-sheets gives insight as to which beta-sheets are parallel and which are anti-parallel. Parallel beta-sheets have "crooked" h-bonding. This makes them more unstable than the anti-parallel beta-sheets which have straight h-bonds between them. Alpha helices are made by hydrogen bonds between the carbonyl of the first and the amino group of the fifth amino acids. The helix follows an i+4 bonding pattern and possesses a right handed turn every ~3.6 amino acids. The biological unit of malate dehydrogenase possesses a total of 12 alpha-helix regions.
Ligand
Citric acid serves as the of malate dehydrogenase.
Hydropathy of Malate Dehydrogenase
To understand why a protein possesses a particular structure it is important to know that protein folding is driven by hydrophobic interactions. One would think that placing a hydrophobic region in water would create more disorder. However, just because something is unfavorable does not mean that it creates more disorder. In fact the opposite is true. When a hydrophobic residue is placed in an aqueous environment, it forces water molecules to interact with one another in such a way that is actually more organized than if hydrophilic residues were in the same position. Less hydrogen bonding interactions are available to a given water molecule. In essence, the concentration of polar groups has been reduced. The structure that is formed by a hydrophobic group in an aqueous environment is called a clathrate. A clathrate resembles a crystalline structure...similar to that of ice, in an aqueous environment with a temperature higher than the melting point. Therefore it is not favorable for the water molecules to be in a more restricted conformation.
The gives vital insight as to why certain residues are buried in the middle of the three dimensional structure and others are not. Hydrophilic residues (blue), are seen more or less on the outer surface of the protein, whereas hydrophobic residues (gray) are seen buried in the middle. more clearly shows the hydrophobic residues.
Solvent Interaction
The hydropathy of a protein provides insight as to how it will interact in the presence of a solvent, however this is more clearly seen when the . Water (blue), interacts with much of the exterior of the protein (white). The only place where water is seen moving into the middle of the three dimensional structure is at the cleft between the two subunits that make up the biological unit, and where the ligand binds the enzyme. The cleft between the two subunits and its interaction with water is more clearly seen .
Important Residues
The residues which are labeled by their charge. Dark blue residues around the ligand have a positive charge, light blue residues around the ligand hold a positive charge in basic conditions and are basic residues. White residues are neutral and carry no charge. The blue residues interact with the electronegative oxygen molecules that are part of the ligand. Perhaps it is easier to see the residues which contact the ligand in a more .
Two of the residues that are in contact with the ligand are also part of the These four residues are an aspartate, arginine,histadine, and alanine.