Sandbox Reserved 896
From Proteopedia
| This Sandbox is Reserved from Jan 13 through July 31, 2020 for use in the course Protein Structure in Drug Discovery taught by Karen C. Glass at the ACPHS, Colchester, United States. This reservation includes Sandbox Reserved 895 through Sandbox Reserved 901. |
To get started:
More help: Help:Editing |
Contents |
Structural and Functional Overview of Human BRD2 (RING3) Protein
|
General Overview
Human bromodomain-containing protein 2 (BRD2) is a highly conserved and ubiquitously expressed protein involved in transcriptional regulation and recognition of post-translational histone modifications. BRD2 is a bromodomain and extra-terminal domain (BET) family protein, which are characterized by the presence of two adjacent bromodomains as well as an extra-terminal domain. Bromodomain and extra terminal domain (BET) family proteins are a group of related proteins involved in the specific recognition of acetylated lysine residues on chromatin and subsequent transcriptional activation [2]. As a BET protein, BRD2 follows this structural motif with its structure consisting of an NET domain at the N-terminus, followed by bromodomain 2 (BRD2-BD2), and finally bromodomain 1 (BRD2-BD1) at the C-terminus. Human BRD2 is encoded by the gene BRD2, which consists of 11 exons that cover more than 6 kb of genomic DNA [1]. This gene was formerly known as really interesting new gene 3 (RING3) but was later renamed to BRD2.
Function
Human BRD2 protein is a Serine-Threonine kinase and chromatin regulator found ubiquitously amongst the nuclear envelope of 223 tissue types. The activity of BRD2 is increased during cell proliferation. BRD2 specifically recognizes an N-acetyl-lysine residue at position 12 of histone H4 [3]. BRD2-BD1 and BD2 preferentially bind diacetylated peptides with optimal spacing between N-acetyl-lysine residues. BD1 has particular affinity for diacetylated lysine residues at position 5 and 8 of H4, whereas BD2 is significantly more promiscuous [6]. It is presumed that one of the functions of this recognition is to prevent deletion or erasure of post-translational histone markers during the mitotic cell cycle. The transcriptional regulation activity of BRD2 is also mediated through its positive regulation of E2F-dependent cell cycle progression (direct stimulation of E2F reporter activity) [1]. As E2F’s central function is to promote the synthesis of proteins needed for G1 to S transition, BRD2-BD1 is directly implicated in the regulation of the cell cycle. BRD2-BD1 also interacts with latency-associated nuclear antigen 1 (LANA-1), a protein involved in Kaposi’s sarcoma-associated herpesvirus [5]. BRD2-BD2 is known to facilitate recruitment of other BET proteins to induce gene expression in specific phases of the cell cycle [6].
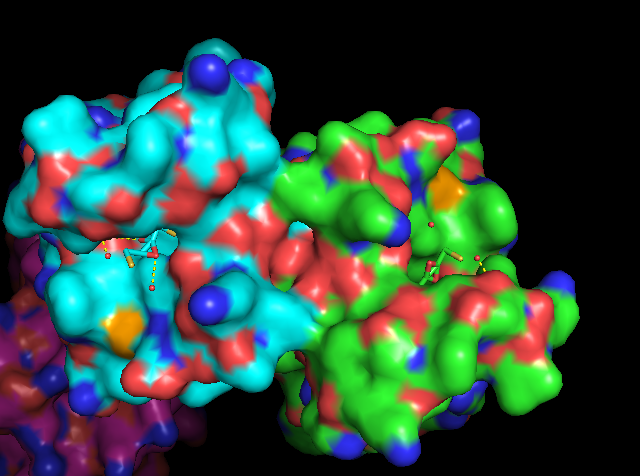
Figure 1: Surface view of BD1 binding N-acetyl-lysine 12 of histone H4. The hypoacetylated side chain of lysine 8 that coordinates the dimer interface cannot be seen in this figure. Figure produced by PyMol (PDB ID: 2DVQ) [17].
Structure
The BRD2 protein consists of three domains: C-terminal bromodomain 1 (BD1), bromodomain 2 (BD2), and N-extra-terminal domain. As BD1 plays the primary role in coordinating the N-acetyl-lysine ligand, this domain’s structure is of the most functional importance.
BD1 Structure
BD1 contains a left-handed alpha-helical bundle formed by four alpha helices: aZ, aA, aB, and aC. It's structure is similar to bromodomains of p300/CBP-associated factor (PCAF), GCN5, and double bromodomain module of TAF1. The active site of BD1 is a deep cleft consisting of a ZA loop (extended long loop which is named so due to connecting the aZ and aA helices) and a BC loop (another loop which connects aB and aC loops). The hydrophobic core is mostly stabilized by conserved hydrophobic residues and few hydrophilic residues.
BD1 vs. BD2 Drug Selectivity
Genetic divergence between the structures of ZA and BC loops in BD1 vs. BD2 is what allows selective drug targeting of one bromodomain In BD1, the shortest communication path between ZA and BC loops is a chain consisting of Leu110 Tyr113 Ile117 Asn151 Tyr153 Asn156 [4]. In BD2, the shortest communication path between ZA and BC loops is a chain consisting of Leu383 Asp385 Ile 389 Tyr428 Asn429c [4]. The precise mechanism for which this selection occurs is unknown, but it is known that Leu383 and Asn429 are the two most important residues of BD2 that allow binding of the RVX-208 inhibitor molecule [4]. ABBC-744 is another preclinical inhibitor that takes advantage of this divergence.
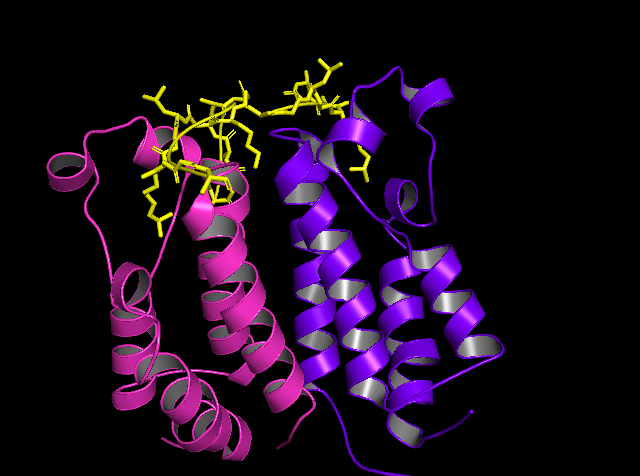
Figure 2: Tertiary Structure of BRD2-BD1 in complex with N-acetyl-lysine 12 of histone H4. Figure produced by PyMol (PDB ID: 2DVQ) [17].
Medical Relevance
Role in Kaposi’s Sarcoma-Associated Herpesvirus
BRD2 interacts with LANA-1, a key protein in Kaposi’s sarcoma-associated herpesvirus. BRD2 facilitates the tethering of LANA-1’s c-terminal domain to host chromatin and is a potential drug target for Kaposi’s sarcoma [6].
Role in Acute Myeloid Leukemia
Recent literature has shown that BD1 is a key domain in the maintenance of malignant phenotypes of acute myeloid leukemia cells. A BD1-specific inhibitor, iBET-BD1 was able to displace chromatin-bound BRD4 in addition to BRD2, indicating that this inhibitor may be effective on the BD1 domain of all BET proteins. This inhibitor’s primary therapeutic outcome were induction of cell cycle arrest and apoptosis, but it also contributed to reduced clonogenic capacity of primary human acute myeloid leukemia cells. Selective BD2 inhibitor RVX-208 demonstrates potent antitumor activity in vitro [4].
Role in Acute and Chronic Lymphoma
Researchers demonstrated a marked increase in BRD2 levels within peripheral blood lymphocytes in both acute and chronic lymphoma patients [1].
Role in Amelioration of Inflammatory Disease
BRD2-BD2 Inhibitor GSK620 shows marked specificity for BD2 over BD1. Similar to its predecessor, iBET-BD2, it shows efficacy among all BET family proteins. GSK620 shows significantly increased oral bioavailability in rodent models. This novel inhibitor of BD2 demonstrates dose-dependent inhibition of progressive cartilaginous destruction, joint swelling, ankylosis, synovitis, cartilage, damage, pannus formation, and bone resorption [6].
Role in Juvenile Myoclonic Epilepsy
Five specific single-nucleotide polymorphisms associated with BRD2 (RING3) gene have been linked to development of juvenile myoclonic epilepsy. The HLA locus on chromosome 6p21) is linked to juvenile mycotic epilepsy, and has been referred to as EJM1. A 2003 study by Pal et al. suggests that this EJM1 is actually the same gene as BRD2 (RING3) [7].
Sequence Alignment of BET Proteins
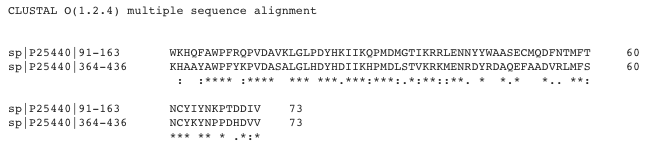
Figure 3: BRD2-BD1 to BRD2-BD2 Alignment
Sequences derived from Uniprot. Figure produced by Clustal Omega [18].
Key: P25440 (91-163) = BRD2-BD1; P25440 (364-463) = BRD2-BD2; * = identical residue; . = chemically similar residue; : = very chemically similar residue
Sequence Identity: 39 / 73 = 53%
Sequence Similarity: (39 + 23) / 73 = 85%
As stated above, it is the low level of sequence identity shared between BRD2-BD1 and BRD2-BD2 that allows for selective drug targeting. The sequence identity is most divergent in the regions consisting the ZA and BC loops, with BD2 containing key residues of Leu383 and Asn429.
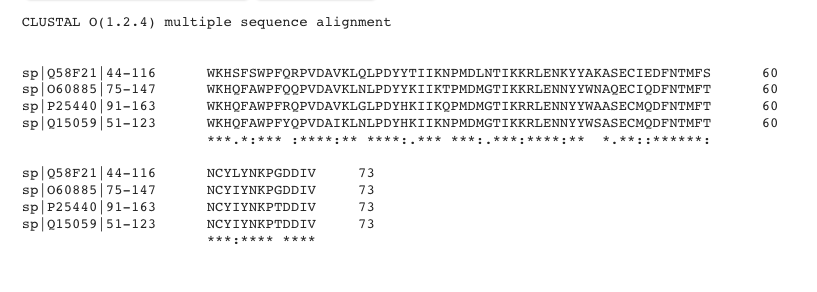
Figure 4: BD1 Alignment Among BET Proteins
Sequences derived from Uniprot. Figure produced by Clustal Omega [18].
Key: P25440 = BRD2; Q15059 = BRD3; O60885 = BRD4; Q58F21 = BRDT; * = identical residue; . = chemically similar residue; : = very chemically similar residue.
Overall Sequence Identity: 50 / 73 = 68%
Overall Sequence Similarity: (50 + 15) / 73 = 89%
Figure 5: BD2 Alignment Among BET Proteins
Sequences derived from Uniprot. Figure produced by Clustal Omega [18].
Key: P25440 = BRD2; Q15059 = BRD3; O60885 = BRD4; Q58F21 = BRDT; * = identical residue; . = chemically similar residue; : = very chemically similar residue.
Overall Sequence Identity: 47 / 73 = 64%
Overall Sequence Similarity: (47 + 17) / 73 = 88%
Figure 6: NET Alignment Among BET Proteins
Sequences derived from Uniprot. Figure produced by Clustal Omega [18].
Key: P25440 = BRD2; Q15059 = BRD3; O60885 = BRD4; Q58F21 = BRDT; * = identical residue; . = chemically similar residue; : = very chemically similar residue.
Overall Sequence Identity: 57 / 83 = 69%
Overall Sequence Similarity: (57 + 17) / 83 = 89%
These findings indicate that BD2 is the most structurally diverse domain within BET family proteins, showing decreased sequence identity and similarity as compared to BD1 and NET domains.
References
[1] Taniguchi Y. The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins. International Journal of Molecular Sciences. 2016Jul;17(11):1849.
[2] Nakamura Y, Umehara T, Nakano K, Jang MK, Shirouzu M, Morita S, et al. Crystal Structure of the Human BRD2 Bromodomain: INSIGHTS INTO DIMERIZATION AND RECOGNITION OF ACETYLATED HISTONE H4. Journal of Biological Chemistry. 2006;282(6):4193–201.
[3] Umehara T, Nakamura Y, Jang MK, Nakano K, Tanaka A, Ozato K, et al. Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain. Journal of Biological Chemistry. 2010Apr;285(10):7610–8.
[4] Wang Q, Li Y, Xu J, Wang Y, Leung EL-H, Liu L, et al. Selective inhibition mechanism of RVX-208 to the second bromodomain of bromo and extraterminal proteins: insight from microsecond molecular dynamics simulations. Scientific Reports. 2017;7(1).
[5] Viejo-Borbolla A, Ottinger M, Bruning E, Burger A, Konig R, Kati E, et al. Brd2/RING3 interacts with a chromatin-binding domain in the Kaposi's Sarcoma-associated herpesvirus latency-associated nuclear antigen 1 (LANA-1) that is required for multiple functions of LANA-1. Journal of Virology. 2005Nov;
[6] Gilan O, Rioja I, Knezevic K, Bell M, Yeung M. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science. 2020Apr24;368(6489):387–94.
[7] Pal DK, Evgrafov OV, Tabares P, Zhang F, Durner M, Greenberg DA. BRD2 (RING3) Is a Probable Major Susceptibility Gene for Common Juvenile Myoclonic Epilepsy. The American Journal of Human Genetics. 2003;73(2):261–70.
[8] UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. Bromodomain-containing protein 2 [Internet]. UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. State Secretariat for Education, Research, and Innovation; 2020 [cited 2020Apr28]. Available from: https://www.uniprot.org/uniprot/P25440#expression
[9] UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. Bromodomain-containing protein 3 [Internet]. UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. State Secretariat for Education, Research, and Innovation; 2020 [cited 2020Apr28]. Available from: https://www.uniprot.org/uniprot/Q15059
[10] UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. Bromodomain-containing protein 4 [Internet]. UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. State Secretariat for Education, Research, and Innovation; 2020 [cited 2020Apr28]. Available from: https://www.uniprot.org/uniprot/O60885
[11] UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. Bromodomain testis-specific protein [Internet]. UniProt ConsortiumEuropean Bioinformatics InstituteProtein Information ResourceSIB Swiss Institute of Bioinformatics. State Secretariat for Education, Research, and Innovation; 2020 [cited 2020Apr28]. Available from: https://www.uniprot.org/uniprot/Q58F21
[12] RCSB Protein Data Bank. 2DVQ: Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide [Internet]. RCSB PDB. [cited 2020Apr28]. Available from: https://www.rcsb.org/structure/2DVQ
[13] Tommaso PD, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang J-M, et al. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Research. 2011Sep;39(suppl).
[14] Armougom F, Moretti S, Poirot O, Audic S, Dumas P, Schaeli B, et al. Expresso: automatic incorporation of structural information in multiple sequence alignments using 3D-Coffee. Nucleic Acids Research. 2006Jan;34(Web Server).
[15] Osullivan O. 3DCoffee: Combining Protein Sequences and Structures within Multiple Sequence Alignments. Journal of Molecular Biology. 2004;
[16] Notredame C, Higgins DG, Heringa J. T-coffee: a novel method for fast and accurate multiple sequence alignment. Thornton J, editor. Journal of Molecular Biology. 2000;302(1):205–17.
[17] The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
[18] 18. Sievers F, Higgins DG. Clustal Omega, Accurate Alignment of Very Large Numbers of Sequences. Methods in Molecular Biology Multiple Sequence Alignment Methods. 2013;:105–16.