From Proteopedia
proteopedia linkproteopedia link DNA Gyrase
| Sandbox reserved 1225. Click above on edit this page to modify. Be careful with the < and > signs.
You may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue.
Function
DNA Gyrase is a protein located mainly in Prokaryotes, specifically bacteria. It falls under the Topoisomerase family and can be referred to as Topoisomerase II. This family is commonly responsible for the protein-protein interactions. DNA Gyrase is responsible for negatively induced supercoiling. This method of supercoiling is used to reduce the strain caused by the twists in DNA. It aids in compaction, as well as separation of DNA. As one can see, it plays a significant role in prokaryotes.
DNA gyrase is a molecular protein responsible in producing negative supercoils to reduce the tension and stress produced from the unwinding activity of helicase. DNA Gyrase uses free ATP produced from hydrolysis to effectively reduce the tension in the DNA. It does so by initiating negative supercoiling in the DNA. This negative supercoiling is the opposite of how DNA winds its self.
Structural highlights
The overall structure of DNA gyrase contains more that 100 base pairs. DNA Gyrase consists of mainly two subunits. This First known as A contains the tyrosine which is responsible for cleavage. The second subunit B is responsible to the binding of ATPase’s active site. Both subunits work together to produce the negative supercoiling produced in DNA strands. The two subunits are often recognized as an A2B2 tetramer. The part of the DNA intended to be cleaved is called the G-segment. DNA gyrase also consists of WHD and TOPRIM, Winged Helix Domain, and Topoisomerase-Primase respectively. The enzyme however is still being researched because of the lack of high resolution gyrase bind to DNA.
Mechanism
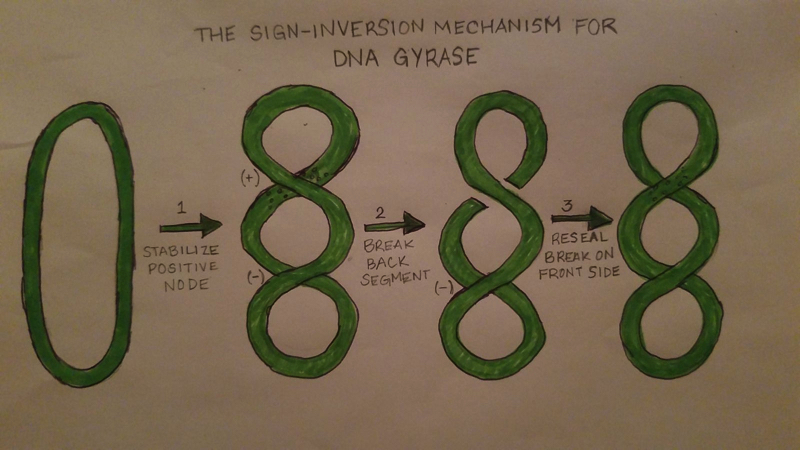
DNA Gyrase works by introducing conformational changes to itself as well as the protein complex. The Gyrase enzyme begins by interacting with the DNA. This causes the formation of the three gates that can be opened and closed. The G-segment, introduced under the “structure” section, forms a bond and binds to the first of the 3 gates, the central DNA gate. This binding of the gate produces chiral wrapping which in then produces a T-segment with in the last of the 3 gates, the N gate. After which, the N-gate binds with ATP which then causes the gate to close. Once this process is completed, the G-segment is released from the last of the 3 gates, the C-gate. This process causes a net result of 2 negative supercoils and a linkage difference of 2 from the initial linkage.
Disease
Researches specifically study this protein for multiple reasons. It could possibly help with antibacterial chemotherapy.
|
References
Basu, Aakash, et al. "Structural Dynamics and Mechanochemical Coupling in DNA Gyrase." Journal of Molecular Biology, vol. 428, no. Part B, 08 May 2016, pp. 1833-1845. EBSCOhost, doi:10.1016/j.jmb.2016.03.016.
Biochemistries. "The Mechanism of Negative DNA Supercoiling A..." Biochemistries. N.p., 23 Feb. 2014. Web. 04 May 2017.
Database, GeneCards Human Gene. "TOP2A Gene(Protein Coding)." GeneCards Is a Searchable, Integrative Database That Provides Comprehensive, User-friendly Information on All Annotated and Predicted Human Genes. N.p., n.d. Web. 04 May 2017
"DNA Gyrase." DNA Gyrase - ScienceDirect Topics. N.p., n.d. Web. 04 May 2017.
Gore, J., Z. Bryant, M. D. Stone, M. Nöllmann, N. R. Cozzarelli, and C. Bustamante. "Mechanochemical Analysis of DNA Gyrase Using Rotor Bead Tracking." Nature. U.S. National Library of Medicine, 05 Jan. 2006. Web. 04 May 2017.
Rahimi, H., Najafi, A., Eslami, H., Negahdari, B., & Moghaddam, M. M. (2016). Identification of novel bacterial DNA gyrase inhibitors: An in silico study. Journal Of Research In Pharmaceutical Sciences, 11(3), 250-258.
Schoeffler, A. J., May, A. P., & Berger, J. M. (2010). A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Research, 38(21), 7830-7844. doi:10.1093/nar/gkq665
Travers, A., & Muskhelishvili, G. (2015). DNA structure and function. FEBS Journal, 282(12), 2279-2295. doi:10.1111/febs.13307
Weigel, L. M., G. J. Anderson, and F. C. Tenover. "DNA Gyrase and Topoisomerase IV Mutations Associated with Fluoroquinolone Resistance in Proteus Mirabilis."Antimicrobial Agents and Chemotherapy. American Society for Microbiology, Aug. 2002. Web. 04 May 2017.
Wendorff, T.J, B.H Schmidt, P. Heslop, C.A Austin, and J.M. Berger. "The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage." J.Mol.Biol. N.p., n.d. Web. 04 May 2017.
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644

