Lignostilbene-α,β-dioxygenase A (LsdA) is a catalytic enzyme found in bacteria to break down cellulose components.
Function
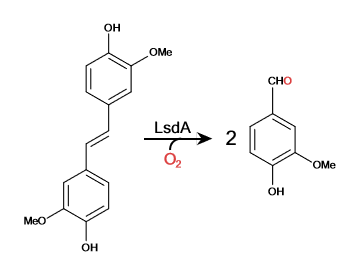
Lignostilbene-α,β-dioxygenase A (LsdA) is an enzyme found in the bacteria Sphingomonas paucimbilis.[1] Lsd is a carotenoid oxygenases that involve the cleavage of carotenoids (CCOs). LsdA is used to catalyze the substrate, lignostilbene, into two vanillin molecules. Vanillin is a member of the benzaldehydes class and plays a role in the plant metabolite[2]. Lignin is found in the plant cell walls, lignocellulose, and it gives the plant its rigid structure. The depolymerization of the lignin can allow for better use of the plants resources as a source of biofuel.
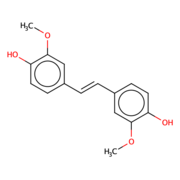
Lignostilbene
The lignostilbene is a compound found in cellulose
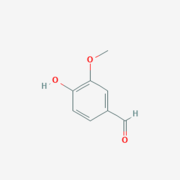
Vanillin
Vanillin is the product from the cleavage of lignostilbene from LsdA.
Cleavage of Lignostilbene
Broader Implications
Lignocellulosic biomasses have been used for biofuels, mainly bio-ethanol. The abundant avaiability of the raw material make the use of biomasses ideal. With the rise of carbon emission, biomasses can be used to substitue the need for carbon based fuel, as long as, the biomass is carbon neutral [3]. Lignostilbene is the second most abundant raw material.
Energy Transformation
When Phe 59 is substituted with His and Tyr 101 is substituted with Phe, the k (app/cat) decreases in activity by 15-10 folds. The Lys 134 substitution of Met inhibits the enzyme activity. Phenylazophenol inhibits the LsdA-catalyzed cleavage of lignostilbene with a competitive and uncompetitive inhibition.
Structural highlights
Lignostilbene is a structure with two identical protein chains working together. The proteins are partially composed of (Blue) and the majority by indicated in red. Together the structure work to form its relationship and interactions within itself and other bio-molecular compounds found in the environment.
A is commonly found in the active site of the protein and binds to the ligand. The triad for LsdA are identified as PHE 59, TYR 101, and LYS 134. The are found within the two chains of the protein structure, accompanied by an Iron ion.
A unique structure of the LsdA is the that is found very commonly in CCOs and helps form a funnel-like active site. A few amino acids that interact with the ligand in the by hydrogen bonding are His 167 in pink, Glu 135 in red, and Phe 308 in indigo. The of the quaternary structure shows the interaction of atoms within the structure and how they fit within each other.
The regions are shown in grey and the polar regions are in magenta. The interactions of hydrophobicity show how the structure interacts with itself as typically hydrophilic regions are found within the structure and hydrophobic regions become the outer most regions of the protein.
The is surrounded by histidines that allow for the Iron ion to be kept in place. The four histidines (HIS 167, HIS 218, HIS 282, and HIS 472) form a tetragonal pyramidal and found in the active site. A cap on the seven-bladed β-propeller helps protect it from the solvent. The ion is delocalized when the hydroxystilbenoid is activated by the enzyme-catalyzed deprotonation of the 4-hydroxy group. It can also form a cation when πelectron density from the scissile double bond redistributes the iron-oxy complex. forming a peroxo-substrate cation intermediate.