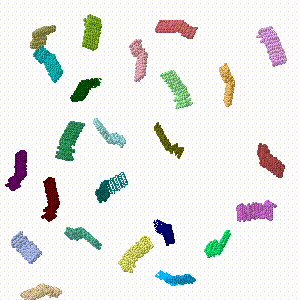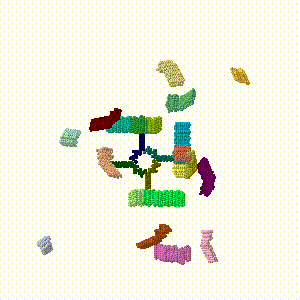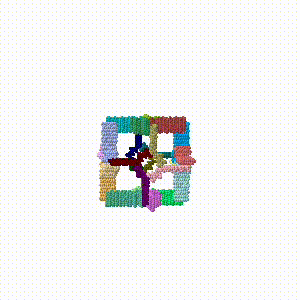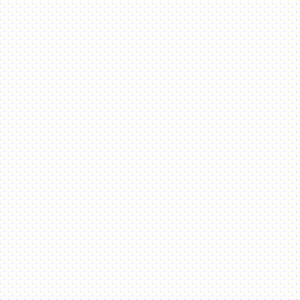In March, 2024, Huddy, Hsia, Kibler, Xu et al. in the team of David Baker (subsequently a Nobel Laureate) published a wide range of synthetic protein nanostructures self-assembled from standardized, engineered alpha-helical protein "building blocks"[1] (). The extensively documented report, in Nature, has 32 authors[1].
The breakthrough here is that instead of designing a single "one-off" desired nanostructure, the Baker group has first designed a series of regular building blocks that can be assembled into diverse nanostructures using straightforward geometric principles. These now enable "construction of protein nanomaterials according to ‘back of an envelope’ architectural blueprints"[1]. There are many potential applications, such as drug delivery or catalysis, which remain to be explored. For example, in 2025 Brunette et al. (Baker lab)[2] reported a multivalent ebola virus vaccine synthetic, self-assembling nanoparticle producing immunity in mice to two kinds of ebola virus. The field of self-assembling protein nanocages was reviewed in December, 2025[3].
Building Blocks
Twistless helix repeat blocks
Click the green links below to change the molecular scene.
Rotate to view the molecule from all sides (drag to rotate).
Zoom with your mouse wheel, or Shift-Drag up/down.
|
In this project, the simplest building blocks consist of anti-parallel alpha helices engineered to be straight and flat, that is twistless helix repeat (THR) protein blocks. A simple example, THR1, is 8g9j, consisting of [4]. Each helix is amphipathic, that is, hydrophobic on the side contacting other helixes, and hydrophilic on the side facing outwards (not shown). The 2.5 Å resolution of 8g9j enabled the modeling of all helix side chains. Non-covalent interactions between helices are nearly all apolar, with a few hydrogen bonds, and two salt bridges (not shown). The , making a highly water soluble building block. The edges of the block are "capped" with charges that prevent these blocks from binding to each other, thus enabling crystallization of this block rather than having it precipitate.
The sequences and binding interfaces of building blocks were designed using Rosetta FastDesign and mainly ProteinMPNN[5][6][7]. Designed sequences were filtered according to likelihood of desired folding and assembly as predicted by AlphaFold2.
33 linear THRs were tested. 23 were solubly expressed. Of 19 characterized by size exclusion chromatography, 13 were primarily monomeric[1].
Five examples of building blocks
Dozens of types of building blocks were designed, synthesized, purified, and their structures and assemblies were determined[1]. [8].
Assemblies
Building blocks were designed with precise angles, and with specific points of attachment between blocks. Most self-assembled into the predicted assemblies. The size of the final assembly can be controlled by the number of helices and their lengths in the building blocks. Examples of assemblies include[9]:
Flat Assemblies
- (120_C3_A_design, branch 60°).
- (90_C4_A_design, 90°).
- (72_C5_A_design, 108°).
- (R20A_design).
- (strut_C6_21_cryo_fit).
- (strut_C6_16_design). Here, the .
Cage Assemblies
Synthetic genes were obtained for 13 nanocage designs; all 13 expressed solubly. Cryo-EM models of seven were symmetric cages resembling the design models[1]. These included:
- (cage_T3_5_+2_design).
- (8TL7, 4.1 Å cryo-EM).
- (cage_O4_32_design).
- (8v2d, 6.8 Å cryo-EM, the initial scene). Here, the .
Inter-Chain Adhesions
In , each chain (block) adheres to three other chains. . The exposed surface of each chain is covered with a mixture of positive + and negative – charges.
Here are four chains in greater detail:
When you hide the three dark colored chains, you will see that they adhere to hydrophobic patches devoid of charges. However, the charges on the edges of these patches may facilitate adhesion.
Animated simulation of self-assembly
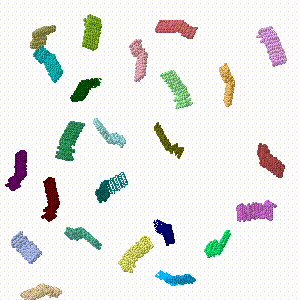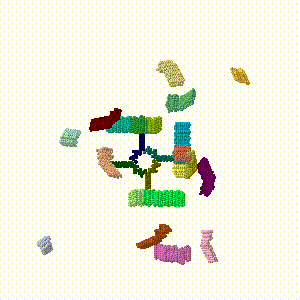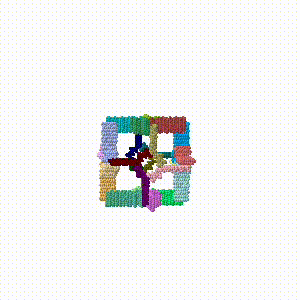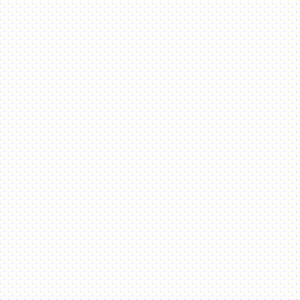
This simulation is crude and very oversimplified ... but heuristic, and hopefully fun to watch.
Alternatively, you can click the link below to get the interactive (rotatable, zoomable, and enlargable) simulation.
Please be patient ... Loading this animation may take up to 30 seconds.
The Toggle Animation button works only for the interactive animation in JSMol.
The popout button  does not work for this animation.
does not work for this animation.
PDB Files
PDB files for the nano structures illustrated above are available in the supplementary materials of Huddy et al., 2024[1], or below. PDB files obtained from Huddy et al., 2024 or derived from those files are re-distributed here under the terms of the original Creative Commons Attribution 4.0 International License.
Method of simulating assembly
The cage model (Cube with no vertices Image:Huddy2024-cube-noverts-cage-o4-32.pdb.gz) has about 272,000 atoms including hydrogens. It was simplified to alpha carbon atoms only (17,256) using the Jmol.jar Java application using Jmol commands "select *.ca; write 0.pdb;". The assembled cage has 24 protein chains.
- In Jmol.jar, 4 chains were dragged out of the assembly to the periphery of the viewport, and each was rotated arbitrarily using the Jmol setting "set picking dragmolecule" (holding down Alt enables rotation). The resulting state was written into a PDB file.
- Repeating this drag/rotate save process six times produced a final model in which none of the protein chains were in their original assembled positions. This resulted in six PDB files 1.pdb, ... 6.pdb.
- The linear morph server by Karsten Theis was used on each pair of PDB files to morph in reverse (towards assembly), producing six morphs 6->5, 5->4, ... 1->0 (where 0 is the fully assembled cube).
- Using a [[Help:Plain text editors plain text editor], the morph files were concatenated in the direction of assembly into a single assembly simulation PDB file.
- The junctions between the six original morph PDB files have one duplicate frame. Those duplicates were removed by text editing.
- An empty (no atoms) MODEL 0/ENDMDL was added to the beginning of the PDB file. This blanks the molecular display between cycles of assembly simulation.
- A morph of the assembled cube rotating slightly was made, and added to the end of the PDB file as the conclusion of the assembly simulation.
- After loading the final assembly simulation PDB file into Jmol, the simulation is played with this command script, which is included in the "Play Assembly Simulation" green link above:
reset;center {-10.484505 11.968994 4.189499}; rotate z -0.04; rotate y 90.22; rotate z -0.42; zoom 112.62;
background white
select all
spacefill 3.0
color chain
anim mode loop 0.5 2.0; # not palindrome
anim on;
anim fps 8; # frames per second
The first line in the above script was copied from the report by Jmol after orienting the cube as desired, and entering the command "show orientation".