User:Anika Zohra Yasin/Sandbox1 3pb3
From Proteopedia
Contents |
Introduction
|
Novel Plasmid-Mediated 16S rRNA methyltransferase also known as NpmA is a plasma-encoded enzyme belonging to the Kanamycin apramycin methyltransferase (Kam) family and consists of structures: 3p2e, 3p2i, 3p2k, 3pb3, and 3mte. NpmA is a methyltransferase or a type of enzyme that methylates one nucleotide in bacterial ribosomes. It specifically methylates on the 16S rRNA at the N1 position of A1408. This methyltransferase activity establishes resistance to aminoglycosides, which function as effective antibiotics against certain types of bacteria.
Bactericidal antibiotics are commonly used to treat diseases caused by both Gram-negative and Gram-positive bacteria. In detail, they promote conformational change by binding to the 30S subunit of the ribosome at the A-site. This results in a loss of fidelity in the protein translation process of the bacteria, and eventually death of the bacteria. However, due to the indiscriminate and increased use, bacteria have evolved and formed resistance to these aminoglycoside antibiotics. The different forms of resistance created against these antibiotics include inactivation by alteration, mutation of the small ribosomal subunit to modify the target site of the antibiotic, or methylation at specific sites on the 16S rRNA to prevent the binding of the antibiotic to the ribosome.
Normally, the antibiotic binds in a tight pocket on the small subunit of the ribosome, disrupting the pairing of codons and anti-codons, to combat the bacteria. However, A clinical isolate of E.Coli has been found to possess, NpmA, an enzyme that flips out the target nucleotide A1408 and adds methyl groups. The specific base is modified in the ribosome, making this pocket unable to fit the antibiotic, and rendering it ineffective against the bacteria. NpmA can be transferred between pathogenic bacteria since it is plasma-encoded. Thus, it poses a threat to the use of aminoglycosides antibiotics in clinical settings.
Structure
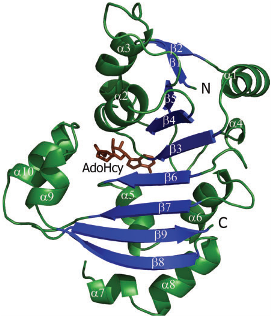
is a 2 chain structure with a 225 amino acid base sequence. Each chain consists of 10 and 9 . The seven-stranded ß sheet (ß5↑–ß4↑–ß3↑–ß6↑–ß7↑–ß9↓–ß8↑) adopts a Class I methyltransferase fold.
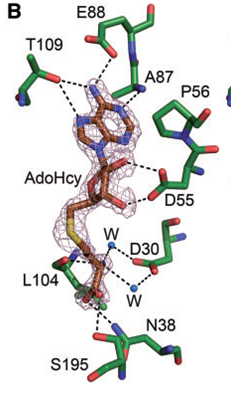
Amino acid residues are conserved in the KAM family. Specific amino acid residues ASP30, TRP107 and TRP197 are crucial to NpmA methyltransferase activity. facilitates cofactor binding by coordinating the carboxypropyl moeity through two water molecules. stabilize the binding, since the target base is stacked between these two residues within the active site of NpmA. Each chain of the protein binds to the ligand, (SAH)(AdoHcy), at binding sites and , respectively. At which, NpmA makes 10 direct hydrogen bonds with AdoHcy.
Sequence
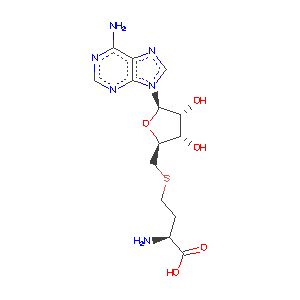
- Ligand Information: S-ADENOSYL-L-HOMOCYSTEINE (SAH) (AdoHcy)
Function
- Methyltransferase Activity
NpmA acts as a methyltransferase also known as methylase which adds a methyl group to specific bases in the ribosome. A binding complex is created between the NpmA and the cofactor, AdoHcy, at certain residues which assists the methylation. The specific residues that outline the cofactor binding site are . NpmA forms 10 direct hydrogen bonds with these residues at this binding site.
ASP30 facilitates cofactor binding by coordinating the carboxypropyl moeity through two water molecules. The target base, Adenosine, is flipped modifying the binding pocket for the aminoglycoside antibiotics. TRP107 and TRP197 stabilize the binding, since this target base is stacked between these two residues within the active site of NpmA. At the N1 position of A1408 in the 16S rRNA, methylation occurs.
- Antibiotic Resistance
Methylation at the N-1 position of A1408 by NpmA will disturb the formation of the hydrogen bond between the A site and aminoglycosides. This highly reduces the binding affinities of aminoglycosides to the target. Kanamycin and apramycin are aminoglycosides affected by this conformational change and since these antibiotics are unable to bind to the ribosomes, they are rendered ineffective.
Significance
Aminoglycosides are the primary agents used to combat life threatening infections caused by gram negative and gram positive bacteria. The ability of these bacteria to confer resistance to these drugs is of serious concern. Research into the method by which these bacteria create this resistance is necessary in order to develop inhibitors. The NpmA structure and knowledge of interactions with its substrate is the first step in developing specific inhibitors of the Kam family of methyltransferases. Once this is acheived, Aminoglycosides that were once rendered ineffective will be able to provide treatment for these infections.
References
1. Amaro, AM. Jerez, CA. Methylation of ribosomal proteins in bacteria: evidence of conserved modification of the eubacterial 50S subunit. (April 1984). <http://jb.asm.org/content/158/1/84.abstract>
2. Husain, N. Obranic, s. Koscinski, L. Seetharaman, J. Babic, F. Bujnicki, M. Maravic, G. and Sivaraman, J. “Structural basis for the methylation of A1408 in 16S rRNA by a panaminoglycoside resistance methyltransferase NpmA from a clinical isolate and analysis of the NpmA interactions with the 30S ribosomal subunit”. (Nov 2010). <http://nar.oxfordjournals.org/content/39/5/1903.full-text-lowres.pdf>
3. Barrett, J. Aminoglycoside. (2002) <http://www.healthline.com/galecontent/aminoglycosides>.
4. “Methyltransferase / Methyltransferases”.(Sino Biological INC.)<http://www.sinobiological.com/Methyltransferase-a-564.html>.
5. “What Is E. Coli?” (Escherichia Coli). (Jun 2011). <http://www.medicalnewstoday.com/articles/68511.php>.
6. Zelinskaya, N. C. Rankin, R. Macmaster, M. Savic, and G. Conn.”Expression, purification and crystallization of adenosine 1408 aminoglycoside-resistance rRNA methyltransferases for structural studies”. (July, 2010). <http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2966526/>.