User:Anique Baig/Sandbox 1
From Proteopedia
Contents |
UDP-sugar 2-Epimerase
The enzyme UDP-sugar 2-epimerase present is in Thermus thermophilus, having a the coding gene WP_011172736. It has a very specific role in catalyzing the last process of the biosynthesis of a rare bacterial sugar UDP-2,3-diacetamido-2,3-dideoxy-D-mannuronic acid (UDP-ManNAc3NAcA). Such sugar has been found in the O-antigens of several pathogenic Gram-negative bacteria, particularly Pseudomonas aeruginosa, Bordetella pertussis and Escherichia albertii and contributes to bacterial virulence and immune evasion.
Reaction Catalyzed by Enzyme
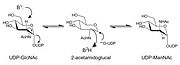
Products of the catalyzed reaction by this 2-epimerase are UDP-ManNAc3NAcA (UDP-2,3-diacetamido-2,3-dideoxy-D-mannuronic acid) formed by the epimerization of C-2 carbon of the sugar ring, which is UDP-GlcNAc3NAcA (UDP-2,3-diacetamido-2,3-dideoxy-D-glucuronic acid). The mechanism exhibited by the enzyme is non-hydrolyzing epimerization process, a feature that is characterized by a method of two steps in terms of chemical reactions. First, UDP anti-elimination and subsequent formation of 2-acetamidoglucal arise. That is followed by a syn-addition of UDP to form the C-2 epimer product.
Such a mechanism is of interest since it is based not on the hydrolysis of the UDP moiety, but on the transient displacement of the latter followed by recombination into another configuration. The allosteric regulation of this enzyme by UDP-GlcNAc was also determined using structural analyses that indicated that an additional, so-called allosteric site exists, binding of which the regulator allosterically activated the enzyme. Notably, the study reports the first structural analysis of a sugar 2-epimerase, acting at UDP-GlcNAc3NAcA as opposed to the more frequently studied UDP-GlcNAc substrates, and gives new insights into the specificity and role of such type of enzymes.
key amino acids of Enzyme
There are important amino acid residues in thermus thermophilus UDP-sugar 2-epimerase whose presence is essential to its catalytic action. These residues are preserved among homologous enzymes and have specific localization in the substrate binding, catalysis as well as stabilization of the intermediate state of a reaction.
Aspartate 98 (Asp 98) is one of the most significant catalytic residues of the enzyme. This residue is placed 3.8 A away from the C-2' carbon ring of the sugar and is regarded as operating in the mechanism of reaction as the general base. It probably operates by abstracting a proton to catalyze the reaction involving anti-elimination of UDP group. Site-directed mutagenesis in which Asp 98 was replaced with asparagine (D98N variant) had a drastic effect in enzyme activity and enabled trapping of the substrate in crystallographic experimentation, thus confirming its catalytic significance.
The other important residue is histidine 208 (His 208) that is close to the substrate and the allosteric activator. It has hydrogen bonding with both C-4 hydroxyl group of UDP-GlcNAc, and a (beta)-phosphoryl oxygen of the UDP-GlcNAc3NAcA substrate. This topology is predictive of a role of His 208 as a general acid, perhaps in the role of proton shuttle mechanism that assists UDP off-loading and re-attachment during turnover.
The Lysine 17 (Lys 17), in turn, helps in substrate binding by hydrogen bonding with the C-6 carboxylate group of the sugar ring. This reaction assists in positioning the substrate accordingly in the active site.
Glutamate 120 (Glu 120) contacts a hydroxyl (C-4 ) of the sugar ring, to place the substrate in the optimal position to be catalyzed. It plays some part in electrostatic interaction that backs the enzyme despite the fact that it is unknown whether it has a specific part to play in transferring protons.
The lysine 132 (Lys 132) has an involvement of hydrogen bonding with C-3 T3 hydroxyl group of the allosteric ligand (UDP-GlcNAc). It has also been seen that it flexes in its conformation according to the different sugar geometries, which could contribute to the efficiency of catalyzing.
The substrate H-bond with a Ser 284 to 2O-phosphoryl residue. Ser 284 is located in the positive face of a helix dipole that probably assists to counteract the negative charges of the pyrophosphorylg group, contributing to the substrate stabilization.
Glutamate 290 (Glu 290) binds to the hydroxyl groups of the ribose of the nucleotide part of the substrate aiding in stabilization of UDP-sugar.
Lastly, Arginine 12 (Arg 12), and Arginine 302 (Arg 302) are also important. Arg 12 is involved in a cation/ |pi| interaction with the nucleotide uracil ring, resulting in the anchoring of the nucleotide base, and at the same time, arg12, in combination with Glutamate 306 (Glu 306), provides structural stabilization through a salt bridge that freezes the active site.
The synergistic combination of these residues forms a very coordinated environment, which allows tightly binding and stabilizing of the substrate, transition states, and speedy catalysis in the 2-epimerase of T. thermophilus.
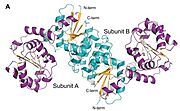
Structure of the Enzyme
UDP-sugar 2-epimerase activity of Thermus thermophilus is well-characterized primary, secondary, tertiary, and quaternary in structure that is essential to its enzymatic activity. High-resolution X-ray crystallography was used to characterize these structural levels in detail in the study.
Primary Structure The major structure of the enzyme is described as primary structure of the enzymes that are the linear amino acid sequence. The complete protein has 360 residues of amino acid. Analysis of sequence alignments of the enzyme to its homologous counterpart(s) in bacteria (Pseudomonas aeruginosa) demonstrated that the enzyme is displaying high sequence identity and similarity (approximately 49% to 60% identity and up to 75 percent similarity) with these homologs; this is particularly so with the enzymes that are related to the biosynthesis of UDP-ManNAc3NAcA. The most important catalytic and binding residues Asp 98, Lys 17, Glu 120, Glu 132, His 208, etc. are extremely well conserved, which suggests that they are critical functional residues.
Secondary Structure The enzyme secondary structure consists of an amalgamation of α-helix and β-sheets arranged into two dominating sections of the structure:
Domain 1 (residues Met 1 to Val 170): Is highly dominated by a seven stranded parallel beta sheets surrounded by six alpha helices.
Domain 2 (Residues Gly 171 to Gly 346): this domain has a parallel 6-stranded beta pleated sheet flanked by nine alpha helices.
There is also a C-terminal α-helix (Ala 350 to Lys 360) which reconnects domain 2 with domain 1 and completes the structure thereby leaving an N- and C-terminus some 13 A apart.
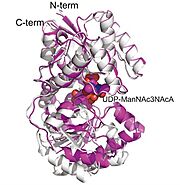
Tertiary Structure The tertiary structure is the general three dimension shapes of a single chain of a polypeptide. The tertiary structure is bilobal with each of the domains bending and interacting with each other. The structural spatial arrangement does help in performing proper orientation of the active and allosteric sites. The active position of a catalytic site was discovered at the cleft between the two domains where it has a chance to move dynamically, since a shift in conformation of 11 A was observed on ligand contact. The active site is encapsulated by specific loops and helices, and contact with the substrate is made much more stable by the presence of conserved residues establishing hydrogen bonds with the substrate and by their electrostatic interactions.
Quaternary Structure Gel-filtration chromatography and X-ray crystallography shows that the enzyme is also a homodimer. The crystal structures are dimers with two subunits in each asymmetric unit with a large area of the dimerization interface burying 3000-4000 A. Dimer interface is mainly hydrophobic with use of both subunits α-helices (most notably His 70-Glu 88, Thr 99-Lys 111, and Pro 131-Ala 142), as well as hydrogen bonds, salt bridges (between Arg 76 and Glu 79), and aromatic stacking interactions (between His 141 residues of each subunit). Dimerization seems significant in regard structural integrity, and possibly also in allosteric regulation.
Methods to analyze the enzyme
Different methods that were used to analyze the enzyme in paper were: X-ray crystallography, Site-directed mutagenesis, Protein expression and purification, Gel filtration chromatography, High performance liquid chromatography, Mass spectrometry, Kinetic analysis
X-ray Crystallography It is a form of central methodology of the paper that is usually covered in a biochemistry course as a basic structural biology procedure. This paper estimates ten high-resolution structures of T. thermophilus 2-epimerase using the X-ray crystallography in various functional states; apoenzyme, substrate and product bound forms. These structures, determined up to 1.8 A resolution, were critical towards learning about the active site structure and the binding of ligands, as well as domain motions during catalysis. Notably, due to this method the researchers were able to visualize the manner in which the enzyme combines its substrate and its allosteric activator, which had not been noticed in this family of enzymes before.
Size-Exclusion Chromatography (Gel-Filtration Chromatography) That method was employed in order to identify the quaternary structure of the enzyme. This enzyme was then filtered through a gel-filtration column and the elution time observed compared with molecular weight markers such as 6-amylase and cytochrome 6-c. The resultant chromatogram demonstrated that T. thermophilus 2-epimerase is a homodimer solution of molecular size of ~82,000 Da. This also aided in corroborating that dimer existed in crystal structures and the dimer is indeed present in a physiological situation and not only in a crystal lattice.
References
Structural analysis of a bacterial UDP-sugar 2-epimerase reveals the active site architecture before and after catalysis Thoden, James B. et al. Journal of Biological Chemistry, Volume 299, Issue 10, 105200